| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
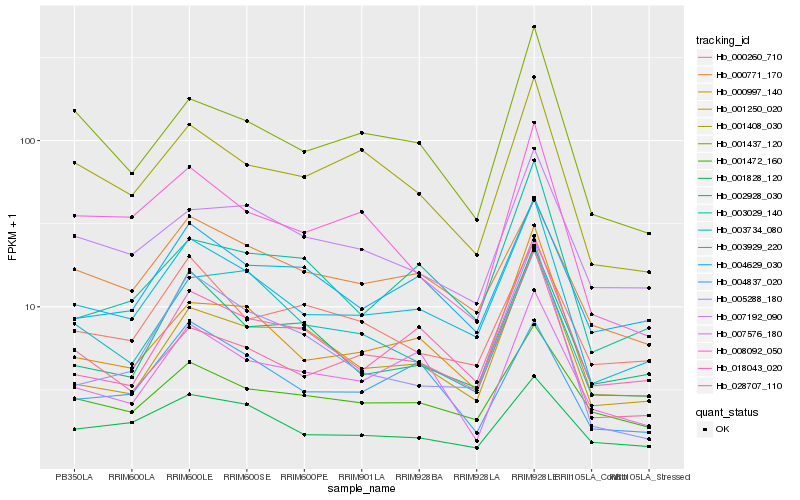
Hb_003929_220 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000997_140 |
0.0980183848 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000771_170 |
0.103741703 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 11, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_028707_110 |
0.106448357 |
- |
- |
PREDICTED: uncharacterized protein LOC105645798 [Jatropha curcas] |
| 5 |
Hb_001408_030 |
0.1085108661 |
- |
- |
PREDICTED: stromal 70 kDa heat shock-related protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001472_160 |
0.1094217954 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 7 |
Hb_001437_120 |
0.1100721525 |
- |
- |
PREDICTED: ruBisCO large subunit-binding protein subunit alpha [Jatropha curcas] |
| 8 |
Hb_004629_030 |
0.111423314 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_018043_020 |
0.1120687904 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007192_090 |
0.1140959371 |
- |
- |
PREDICTED: elongation factor Tu, chloroplastic-like [Glycine max] |
| 11 |
Hb_002928_030 |
0.1141445922 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 12 |
Hb_001828_120 |
0.1148146891 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 13 |
Hb_001250_020 |
0.1176797811 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000260_710 |
0.1195858787 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 15 |
Hb_008092_050 |
0.1208455231 |
- |
- |
PREDICTED: uncharacterized protein LOC105142434 [Populus euphratica] |
| 16 |
Hb_003029_140 |
0.1225406753 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007576_180 |
0.1228169115 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 18 |
Hb_004837_020 |
0.1270354775 |
- |
- |
PREDICTED: uncharacterized protein LOC105648271 [Jatropha curcas] |
| 19 |
Hb_003734_080 |
0.1273201599 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 20 |
Hb_005288_180 |
0.1279763483 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |