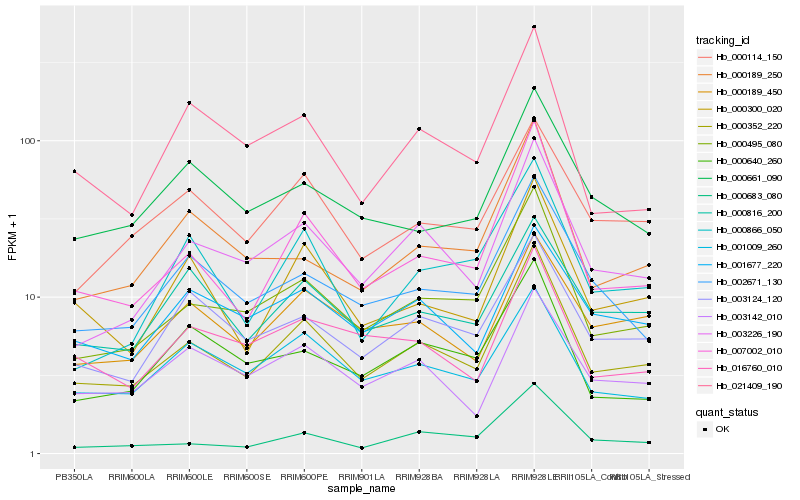
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_220 |
0.0 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000495_080 |
0.0728063772 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 3 |
Hb_007002_010 |
0.0821632581 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 4 |
Hb_003226_190 |
0.0876065087 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 5 |
Hb_000300_020 |
0.0993910814 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial [Jatropha curcas] |
| 6 |
Hb_002671_130 |
0.1145175624 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 7 |
Hb_000114_150 |
0.1152316151 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_003124_120 |
0.1157423296 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 9 |
Hb_001009_260 |
0.117236004 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_021409_190 |
0.1192877701 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 11 |
Hb_000189_450 |
0.1195564741 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000683_080 |
0.1204471895 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 13 |
Hb_003142_010 |
0.1207959532 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 14 |
Hb_000816_200 |
0.1212093902 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 15 |
Hb_016760_010 |
0.121882864 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_000189_250 |
0.1222802368 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000866_050 |
0.1234710639 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 18 |
Hb_001677_220 |
0.1255143629 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 19 |
Hb_000640_260 |
0.1264925286 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000661_090 |
0.1267988523 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |