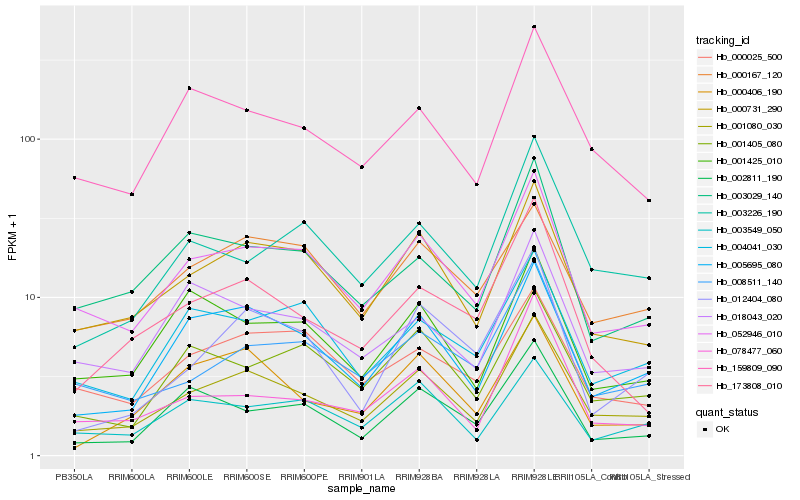
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_030 |
0.0 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 2 |
Hb_052946_010 |
0.0844650601 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 3 |
Hb_000731_290 |
0.0872405392 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_005695_080 |
0.0964951383 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 5 |
Hb_159809_090 |
0.1114789849 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 6 |
Hb_008511_140 |
0.1184454729 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 7 |
Hb_003226_190 |
0.1244278233 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 8 |
Hb_003029_140 |
0.1261239473 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 9 |
Hb_018043_020 |
0.1286487053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001405_080 |
0.1294349271 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 11 |
Hb_000406_190 |
0.129719077 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_173808_010 |
0.1316736276 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 13 |
Hb_078477_060 |
0.1317774222 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 14 |
Hb_004041_030 |
0.133581048 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 15 |
Hb_003549_050 |
0.134136806 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 16 |
Hb_000025_500 |
0.134156793 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 17 |
Hb_001425_010 |
0.1341836222 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 18 |
Hb_002811_190 |
0.1375086809 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 19 |
Hb_000167_120 |
0.1378517603 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 20 |
Hb_012404_080 |
0.1395101567 |
- |
- |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |