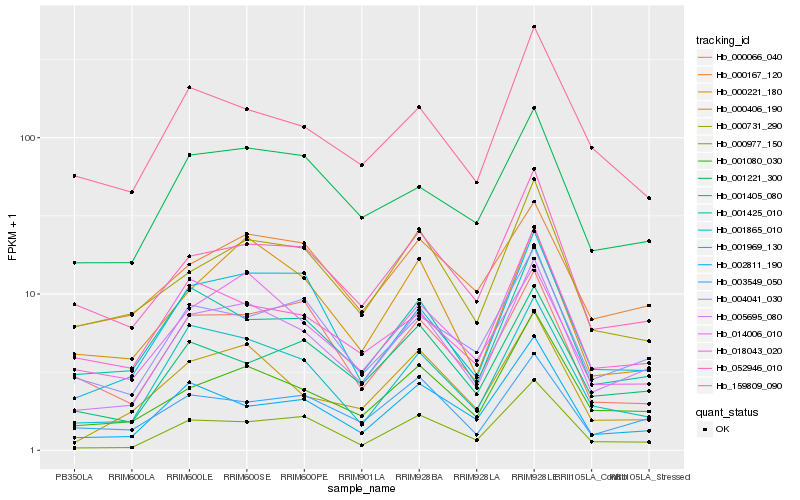
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005695_080 |
0.0 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 2 |
Hb_001080_030 |
0.0964951383 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 3 |
Hb_001865_010 |
0.1039496324 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 4 |
Hb_000406_190 |
0.1051354539 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_001425_010 |
0.1108388025 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 6 |
Hb_001969_130 |
0.1120592256 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 7 |
Hb_052946_010 |
0.1136667663 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 8 |
Hb_004041_030 |
0.1147814522 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 9 |
Hb_001221_300 |
0.115168437 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000731_290 |
0.1163365745 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_003549_050 |
0.1185259969 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 12 |
Hb_001405_080 |
0.119465179 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 13 |
Hb_000977_150 |
0.1223364548 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 14 |
Hb_014006_010 |
0.1244636907 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 15 |
Hb_018043_020 |
0.1250234752 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein MRL1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002811_190 |
0.1250578008 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 17 |
Hb_000221_180 |
0.1265171341 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000167_120 |
0.1303770558 |
- |
- |
PREDICTED: U-box domain-containing protein 14 [Jatropha curcas] |
| 19 |
Hb_000066_040 |
0.1324256087 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 20 |
Hb_159809_090 |
0.132868069 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |