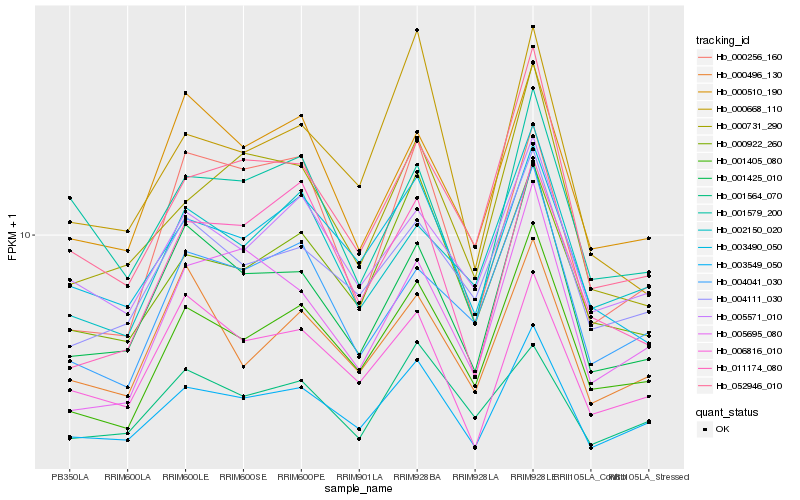
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003549_050 |
0.0 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 2 |
Hb_001405_080 |
0.0858215161 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 3 |
Hb_001425_010 |
0.0971312351 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 4 |
Hb_000922_260 |
0.1002413964 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 5 |
Hb_052946_010 |
0.1038627222 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 6 |
Hb_004111_030 |
0.1048604505 |
- |
- |
PREDICTED: DNA gyrase subunit B, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_006816_010 |
0.1100997992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000731_290 |
0.1102501797 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_003490_050 |
0.1119821761 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005571_010 |
0.1139497599 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 11 |
Hb_001564_070 |
0.1177862086 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 12 |
Hb_005695_080 |
0.1185259969 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 13 |
Hb_004041_030 |
0.1227959064 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 14 |
Hb_000510_190 |
0.1229950652 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 15 |
Hb_002150_020 |
0.1237302618 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_000496_130 |
0.1249770071 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 17 |
Hb_001579_200 |
0.1253461928 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 18 |
Hb_000256_160 |
0.1263256293 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 19 |
Hb_000668_110 |
0.1268862294 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 20 |
Hb_011174_080 |
0.1284228503 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |