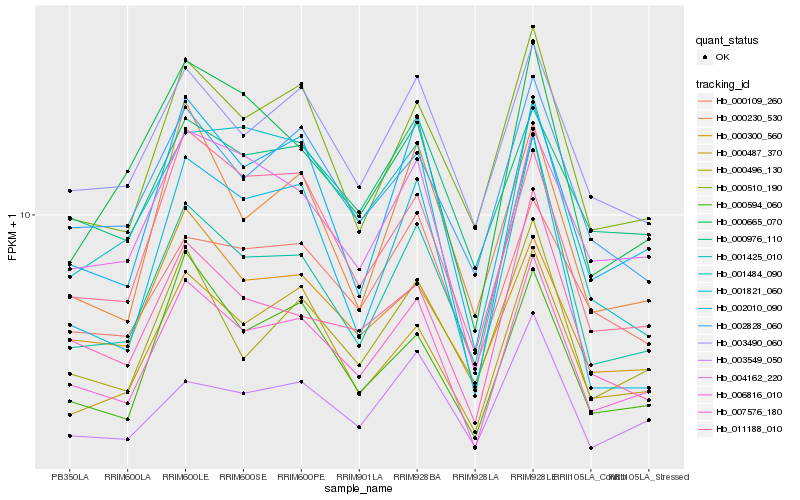
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002010_090 |
0.0834041945 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000510_190 |
0.0976149565 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 4 |
Hb_004162_220 |
0.0981728807 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000487_370 |
0.0992723883 |
- |
- |
- |
| 6 |
Hb_007576_180 |
0.1043905376 |
- |
- |
PREDICTED: probable protein phosphatase 2C 62 isoform X5 [Jatropha curcas] |
| 7 |
Hb_003490_060 |
0.1055955994 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 8 |
Hb_011188_010 |
0.1068734287 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000594_060 |
0.1069513859 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 10 |
Hb_001425_010 |
0.1076756401 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_000496_130 |
0.1085177333 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 12 |
Hb_003549_050 |
0.1100997992 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2-like [Jatropha curcas] |
| 13 |
Hb_002828_060 |
0.1103047821 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000230_530 |
0.1136247175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001484_090 |
0.114493468 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000109_260 |
0.1145369323 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000300_560 |
0.116936998 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000665_070 |
0.1176220399 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001821_060 |
0.120423053 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 20 |
Hb_000976_110 |
0.1205474035 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |