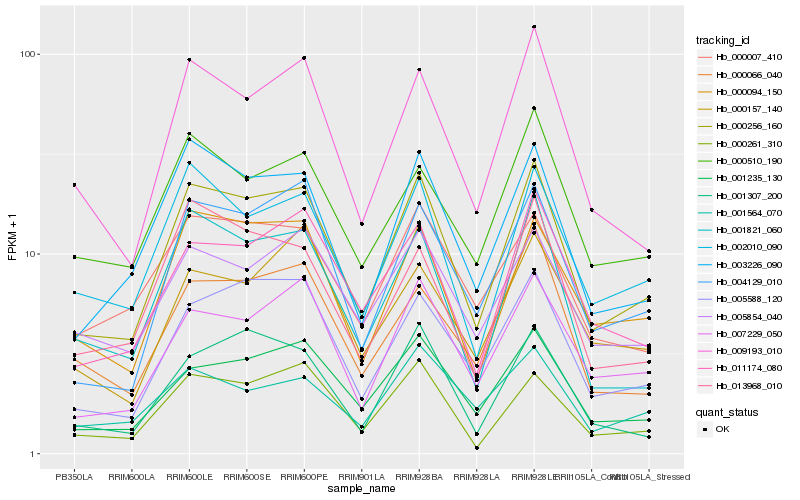
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_160 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 2 |
Hb_001235_130 |
0.0829717197 |
- |
- |
- |
| 3 |
Hb_003226_090 |
0.083414535 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 4 |
Hb_000094_150 |
0.0868624422 |
- |
- |
- |
| 5 |
Hb_004129_010 |
0.0881178143 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 6 |
Hb_005588_120 |
0.0884051492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002010_090 |
0.0919322094 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_007229_050 |
0.0920205024 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000261_310 |
0.0935358585 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 10 |
Hb_009193_010 |
0.0958640705 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 11 |
Hb_011174_080 |
0.0973053088 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 12 |
Hb_001821_060 |
0.0981888276 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 13 |
Hb_005854_040 |
0.1058116005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000007_410 |
0.110133341 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 isoform X2 [Populus euphratica] |
| 15 |
Hb_000510_190 |
0.1112647578 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 16 |
Hb_013968_010 |
0.1142226108 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 17 |
Hb_000066_040 |
0.1143527369 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 18 |
Hb_001307_200 |
0.1146975884 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 19 |
Hb_001564_070 |
0.1190833072 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 20 |
Hb_000157_140 |
0.1196929739 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |