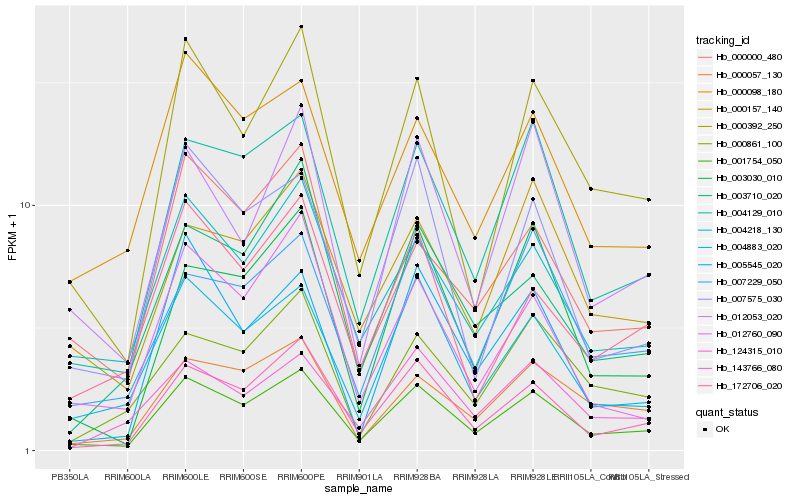
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001754_050 |
0.0 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_143766_080 |
0.0740815858 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004129_010 |
0.0775152935 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 4 |
Hb_000392_250 |
0.0883377749 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 5 |
Hb_004883_020 |
0.0973317976 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 6 |
Hb_000057_130 |
0.1083030893 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 7 |
Hb_004218_130 |
0.1085994471 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 8 |
Hb_007229_050 |
0.1137245632 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007575_030 |
0.1138323736 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 10 |
Hb_003030_010 |
0.1148766159 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 11 |
Hb_012053_020 |
0.117675046 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000098_180 |
0.1194514855 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 13 |
Hb_012760_090 |
0.1224201303 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 14 |
Hb_172706_020 |
0.1233662395 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 15 |
Hb_005545_020 |
0.1251152975 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 16 |
Hb_000000_480 |
0.125645229 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000157_140 |
0.1256505016 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 18 |
Hb_000861_100 |
0.126380468 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 19 |
Hb_124315_010 |
0.1283469842 |
- |
- |
PREDICTED: uncharacterized protein LOC105650413 [Jatropha curcas] |
| 20 |
Hb_003710_020 |
0.128511856 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |