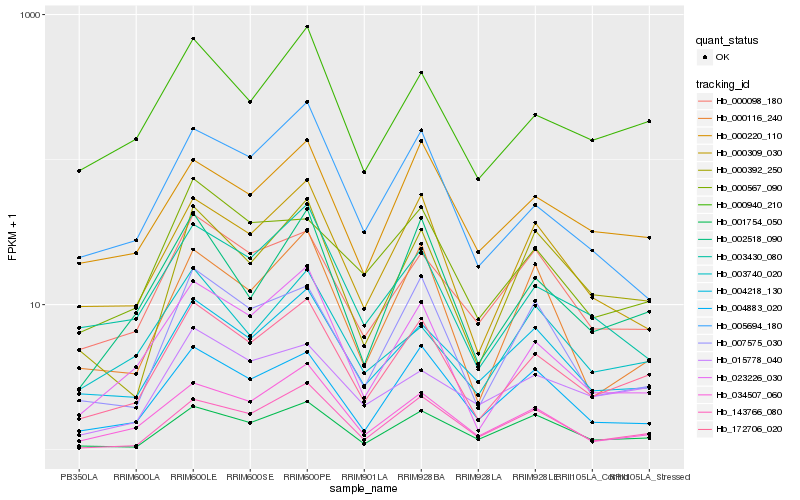
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172706_020 |
0.0 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 2 |
Hb_000940_210 |
0.0955535269 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 3 |
Hb_023226_030 |
0.1029403362 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 4 |
Hb_002518_090 |
0.1076039583 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 5 |
Hb_034507_060 |
0.1104367638 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 6 |
Hb_004218_130 |
0.113076849 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 7 |
Hb_000309_030 |
0.1137207539 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000220_110 |
0.1205510714 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 9 |
Hb_000392_250 |
0.1211832978 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 10 |
Hb_004883_020 |
0.1223851373 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 11 |
Hb_003430_080 |
0.1233449712 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001754_050 |
0.1233662395 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_143766_080 |
0.1264286732 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000098_180 |
0.1270300666 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007575_030 |
0.1294700804 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 16 |
Hb_000116_240 |
0.1362343546 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_005694_180 |
0.136984237 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 18 |
Hb_000567_090 |
0.1375556029 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_003740_020 |
0.1388886221 |
- |
- |
PREDICTED: methylglutaconyl-CoA hydratase, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_015778_040 |
0.1393119635 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |