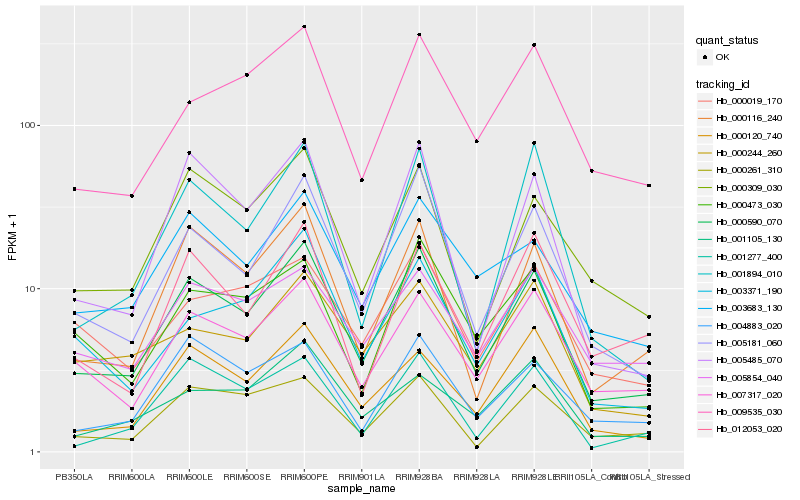
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000590_070 |
0.0 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 2 |
Hb_005181_060 |
0.0794556914 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 3 |
Hb_000116_240 |
0.0933490316 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 4 |
Hb_007317_020 |
0.1041046086 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000309_030 |
0.1046582771 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_005485_070 |
0.1080231715 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000120_740 |
0.1142129561 |
desease resistance |
Gene Name: VARLMGL |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003683_130 |
0.1156574988 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000261_310 |
0.1202452221 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X3 [Jatropha curcas] |
| 10 |
Hb_004883_020 |
0.1208484939 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 11 |
Hb_009535_030 |
0.1226855616 |
- |
- |
CP2 [Hevea brasiliensis] |
| 12 |
Hb_000473_030 |
0.1243282398 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 13 |
Hb_001894_010 |
0.1247720795 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 14 |
Hb_003371_190 |
0.1262681689 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 15 |
Hb_001277_400 |
0.1270280274 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 16 |
Hb_000244_260 |
0.1296839008 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 17 |
Hb_012053_020 |
0.1305892734 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000019_170 |
0.1312580449 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 19 |
Hb_005854_040 |
0.1312867575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001105_130 |
0.1316497788 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |