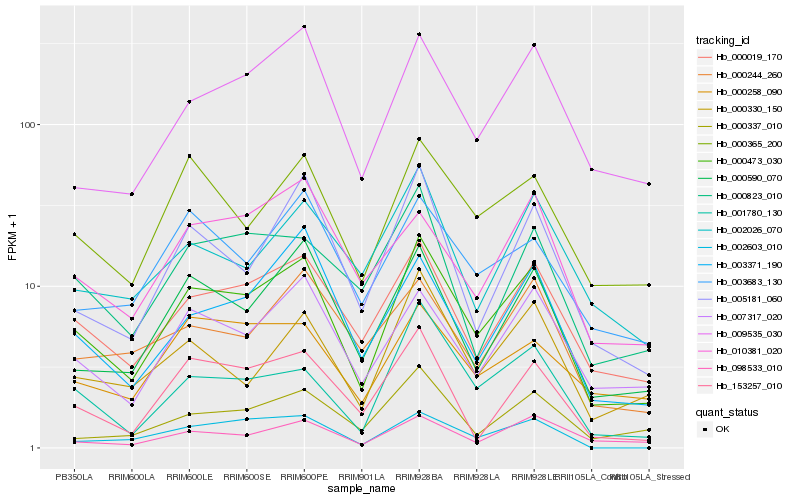
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000473_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 2 |
Hb_000019_170 |
0.1061892857 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 3 |
Hb_000590_070 |
0.1243282398 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 4 |
Hb_007317_020 |
0.1269791724 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000365_200 |
0.1300638715 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 6 |
Hb_009535_030 |
0.134991809 |
- |
- |
CP2 [Hevea brasiliensis] |
| 7 |
Hb_005181_060 |
0.1358957607 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 8 |
Hb_001780_130 |
0.1390542217 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 9 |
Hb_003371_190 |
0.1422130006 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 10 |
Hb_000330_150 |
0.1425686265 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |
| 11 |
Hb_098533_010 |
0.1446541787 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 12 |
Hb_153257_010 |
0.14933724 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000823_010 |
0.1494831305 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 14 |
Hb_000337_010 |
0.1511045606 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002026_070 |
0.1516000114 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 16 |
Hb_003683_130 |
0.1518526336 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002603_010 |
0.151976524 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000258_090 |
0.153025508 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000244_260 |
0.1533639523 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 20 |
Hb_010381_020 |
0.154204191 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |