| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
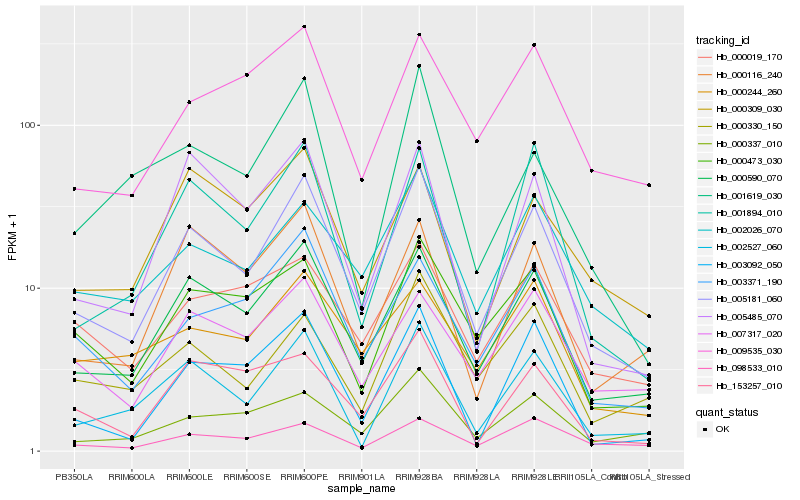
Hb_005181_060 |
0.0 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 2 |
Hb_000590_070 |
0.0794556914 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 3 |
Hb_002527_060 |
0.1249401734 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 4 |
Hb_002026_070 |
0.1280977012 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 5 |
Hb_001894_010 |
0.1306127506 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 6 |
Hb_005485_070 |
0.1328725151 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000473_030 |
0.1358957607 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 8 |
Hb_003092_050 |
0.1381378841 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 9 |
Hb_003371_190 |
0.1391419227 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 10 |
Hb_000337_010 |
0.142410804 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000116_240 |
0.1433600989 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 12 |
Hb_007317_020 |
0.1433646118 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_098533_010 |
0.1458751543 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 14 |
Hb_153257_010 |
0.1492903137 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 15 |
Hb_000019_170 |
0.1509961718 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 16 |
Hb_009535_030 |
0.151129196 |
- |
- |
CP2 [Hevea brasiliensis] |
| 17 |
Hb_000330_150 |
0.1513392629 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |
| 18 |
Hb_000244_260 |
0.1518047287 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000309_030 |
0.1525922883 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001619_030 |
0.1558901066 |
- |
- |
annexin, putative [Ricinus communis] |