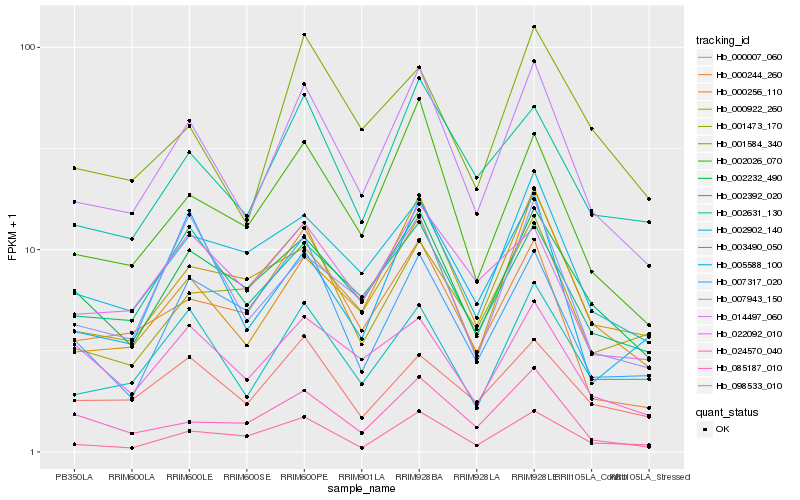
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014497_060 |
0.0 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 2 |
Hb_000256_110 |
0.0912409267 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002026_070 |
0.0954407542 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 4 |
Hb_002902_140 |
0.1061739128 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 5 |
Hb_002232_490 |
0.1076396276 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002631_130 |
0.1108039085 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 7 |
Hb_000922_260 |
0.1148511786 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 8 |
Hb_003490_050 |
0.1153981138 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002392_020 |
0.1185856553 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 10 |
Hb_000007_060 |
0.1211848847 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007943_150 |
0.1221537126 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000244_260 |
0.1235507332 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 13 |
Hb_098533_010 |
0.125082803 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 14 |
Hb_001584_340 |
0.127792998 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 15 |
Hb_085187_010 |
0.1316001237 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 16 |
Hb_022092_010 |
0.1322133056 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 17 |
Hb_007317_020 |
0.1344166313 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_024570_040 |
0.137611473 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 19 |
Hb_005588_100 |
0.1388947815 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_001473_170 |
0.1391217965 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |