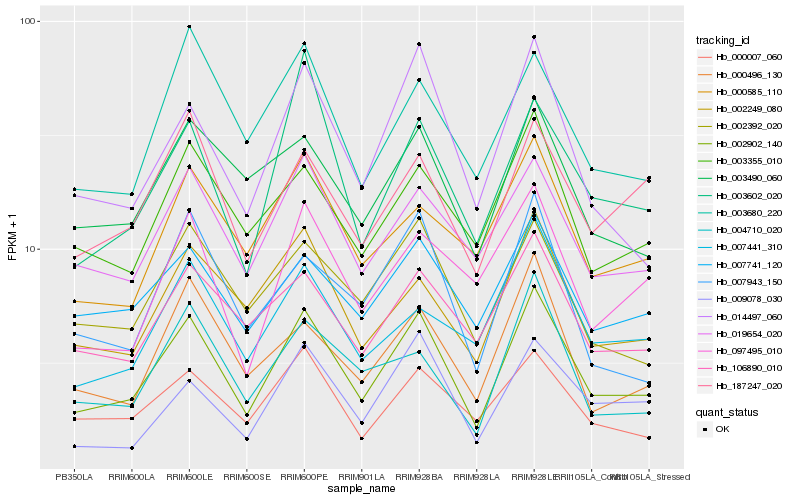
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002902_140 |
0.0 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 2 |
Hb_014497_060 |
0.1061739128 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 3 |
Hb_019654_020 |
0.1078369168 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 4 |
Hb_002249_080 |
0.1099450029 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_000007_060 |
0.1099830371 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_106890_010 |
0.1126422061 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_003355_010 |
0.1134850577 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 8 |
Hb_002392_020 |
0.1135008015 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase [Jatropha curcas] |
| 9 |
Hb_000496_130 |
0.1148039606 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 10 |
Hb_007741_120 |
0.1151175814 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003490_060 |
0.1168293442 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 12 |
Hb_009078_030 |
0.1186614563 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |
| 13 |
Hb_187247_020 |
0.1199006058 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 14 |
Hb_007943_150 |
0.1202000945 |
- |
- |
PREDICTED: proteinaceous RNase P 1, chloroplastic/mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_004710_020 |
0.1203562475 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_000585_110 |
0.1220373982 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 17 |
Hb_097495_010 |
0.1223855047 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003680_220 |
0.1232823543 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 19 |
Hb_007441_310 |
0.1247021126 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003602_020 |
0.1258297576 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial-like [Jatropha curcas] |