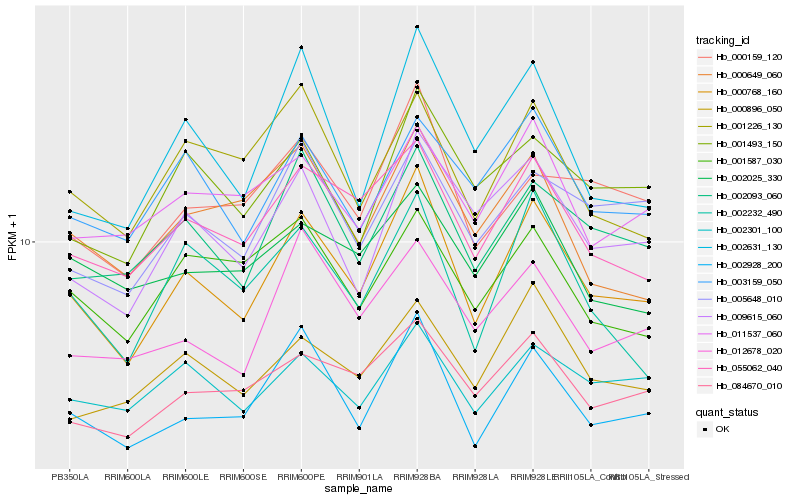
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000768_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 2 |
Hb_002928_200 |
0.0877371338 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 3 |
Hb_055062_040 |
0.0963070473 |
- |
- |
ceramidase, putative [Ricinus communis] |
| 4 |
Hb_002093_060 |
0.0989226957 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_009615_060 |
0.1001227902 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 6 |
Hb_002631_130 |
0.1009431456 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 7 |
Hb_084670_010 |
0.1037657332 |
- |
- |
PREDICTED: thiamine biosynthetic bifunctional enzyme TH1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_002232_490 |
0.1097558137 |
- |
- |
PREDICTED: probable sphingolipid transporter spinster homolog 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000896_050 |
0.1102990539 |
- |
- |
PREDICTED: uncharacterized protein LOC105637668 [Jatropha curcas] |
| 10 |
Hb_000159_120 |
0.1110661834 |
- |
- |
PREDICTED: uncharacterized protein LOC105632857 [Jatropha curcas] |
| 11 |
Hb_002301_100 |
0.1127540423 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 12 |
Hb_001493_150 |
0.1127549748 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 13 |
Hb_005648_010 |
0.1129499766 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 14 |
Hb_012678_020 |
0.1132342342 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003159_050 |
0.1141910245 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_001226_130 |
0.1159190968 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 17 |
Hb_000649_060 |
0.1193861048 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 18 |
Hb_011537_060 |
0.1212469565 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 19 |
Hb_001587_030 |
0.1228129258 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 20 |
Hb_002025_330 |
0.1233774228 |
- |
- |
peptide transporter, putative [Ricinus communis] |