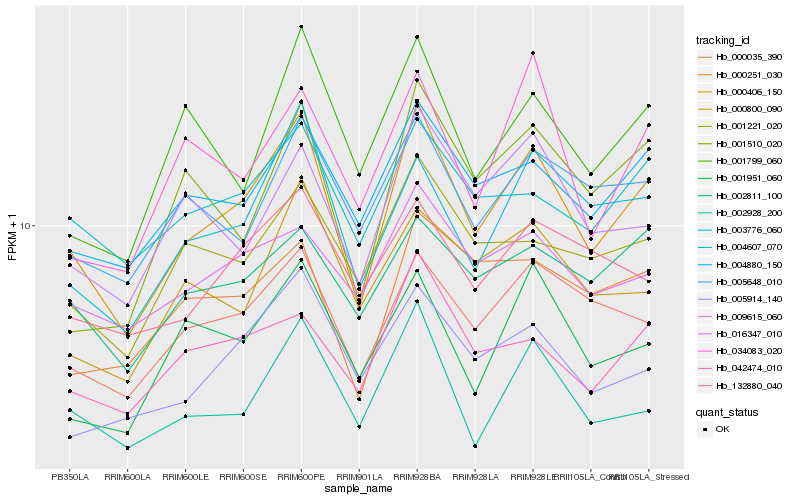
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000406_150 |
0.0 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 2 |
Hb_000035_390 |
0.1068443796 |
- |
- |
hypothetical protein CISIN_1g003355mg [Citrus sinensis] |
| 3 |
Hb_004607_070 |
0.108246237 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 4 |
Hb_004880_150 |
0.1103171243 |
- |
- |
PREDICTED: uncharacterized protein LOC105632834 [Jatropha curcas] |
| 5 |
Hb_002928_200 |
0.1145896821 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 6 |
Hb_042474_010 |
0.1174506316 |
- |
- |
PREDICTED: adenylyltransferase and sulfurtransferase MOCS3 [Jatropha curcas] |
| 7 |
Hb_003776_060 |
0.1208923456 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 8 |
Hb_009615_060 |
0.1210948463 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 9 |
Hb_005648_010 |
0.1212272389 |
- |
- |
PREDICTED: malate dehydrogenase, glyoxysomal isoform X1 [Jatropha curcas] |
| 10 |
Hb_005914_140 |
0.121904593 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 11 |
Hb_001221_020 |
0.1222827391 |
- |
- |
PREDICTED: probable aspartyl aminopeptidase [Jatropha curcas] |
| 12 |
Hb_001799_060 |
0.1231751356 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 13 |
Hb_001951_060 |
0.1257225922 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 14 |
Hb_002811_100 |
0.1257835688 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 15 |
Hb_016347_010 |
0.1260186951 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_001510_020 |
0.1277736523 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 17 |
Hb_000800_090 |
0.1284546314 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000251_030 |
0.128610749 |
- |
- |
nicotinate phosphoribosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_034083_020 |
0.1286502942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_132880_040 |
0.1293246679 |
- |
- |
PREDICTED: protein SUPPRESSOR OF K(+) TRANSPORT GROWTH DEFECT 1-like isoform X2 [Jatropha curcas] |