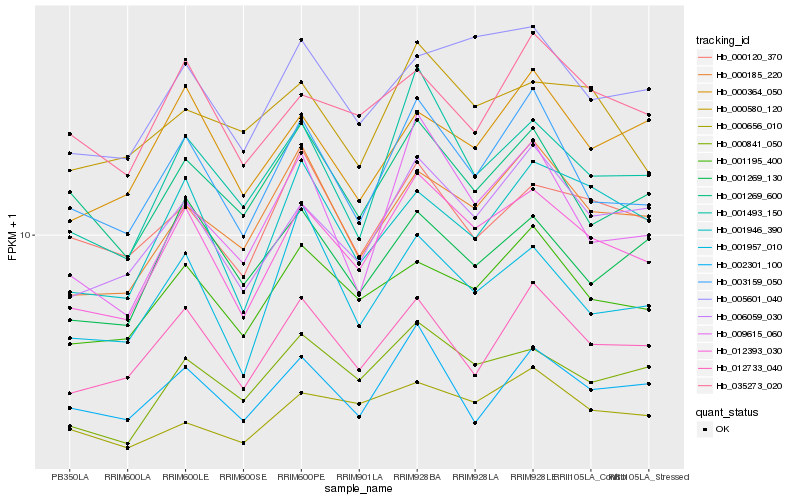
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012393_030 |
0.0 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 2 |
Hb_006059_030 |
0.0677689966 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000841_050 |
0.0725320064 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 4 |
Hb_003159_050 |
0.0745767895 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_001195_400 |
0.075564033 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 6 |
Hb_001493_150 |
0.077906765 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 7 |
Hb_005601_040 |
0.0813114467 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 8 |
Hb_012733_040 |
0.0824689109 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 9 |
Hb_000185_220 |
0.0865646332 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 10 |
Hb_001946_390 |
0.0873004268 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 11 |
Hb_001957_010 |
0.0937288143 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001269_130 |
0.0943196888 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 13 |
Hb_035273_020 |
0.0955440643 |
- |
- |
PREDICTED: amidase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002301_100 |
0.0956614504 |
- |
- |
hypothetical protein JCGZ_21479 [Jatropha curcas] |
| 15 |
Hb_009615_060 |
0.0956673824 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 16 |
Hb_001269_600 |
0.0965756561 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_000364_050 |
0.0978489939 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000580_120 |
0.0981118805 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000656_010 |
0.0989391822 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000120_370 |
0.0991173445 |
- |
- |
type 2 diacylglycerol acyltransferase [Ricinus communis] |