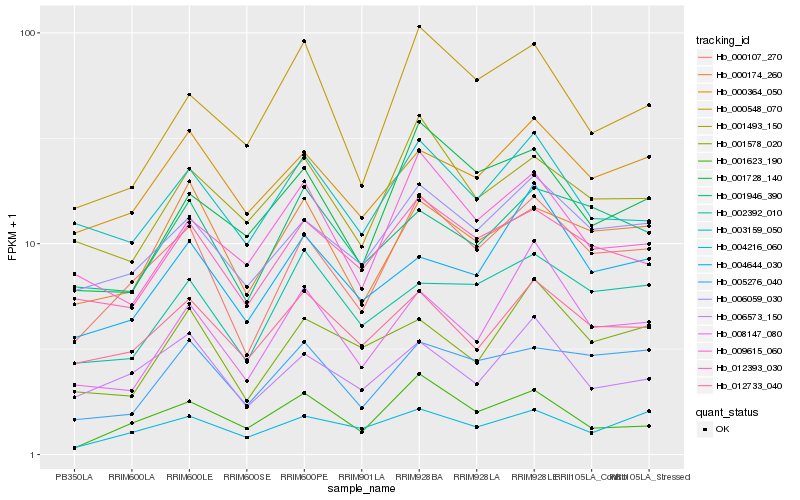
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_270 |
0.0 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_006059_030 |
0.0827216487 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_012393_030 |
0.1040211585 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 4 |
Hb_000174_260 |
0.1149311579 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 5 |
Hb_000364_050 |
0.1161621583 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_012733_040 |
0.1174119772 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 7 |
Hb_005276_040 |
0.1184922816 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 8 |
Hb_006573_150 |
0.1190448535 |
- |
- |
PREDICTED: DNA-directed primase/polymerase protein [Jatropha curcas] |
| 9 |
Hb_003159_050 |
0.1205041879 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_004216_060 |
0.1215641459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001946_390 |
0.1250143457 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 12 |
Hb_001493_150 |
0.1254137273 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 13 |
Hb_002392_010 |
0.1259761412 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 14 |
Hb_001728_140 |
0.1273344717 |
- |
- |
- |
| 15 |
Hb_008147_080 |
0.1275375641 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001578_020 |
0.1291154648 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 17 |
Hb_001623_190 |
0.1297963199 |
- |
- |
PREDICTED: uncharacterized protein LOC105638473 [Jatropha curcas] |
| 18 |
Hb_009615_060 |
0.1298922418 |
- |
- |
PREDICTED: prolyl endopeptidase-like [Jatropha curcas] |
| 19 |
Hb_000548_070 |
0.1299499766 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 20 |
Hb_004644_030 |
0.1305639668 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |