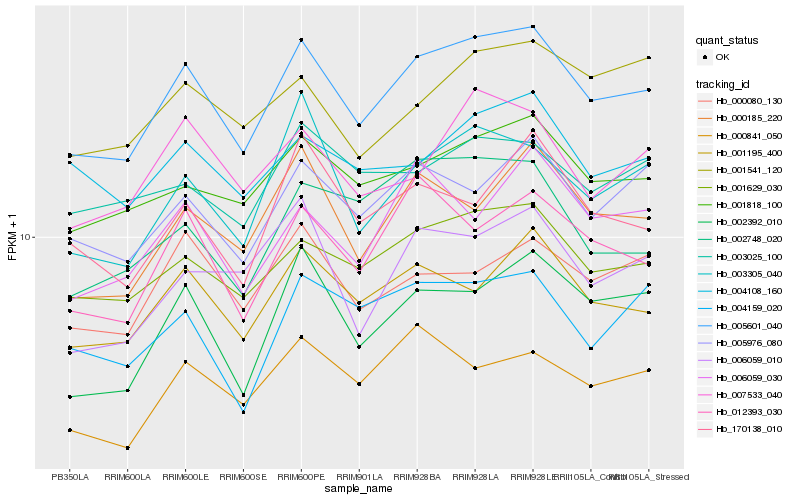
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005601_040 |
0.0 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 2 |
Hb_001629_030 |
0.0593766486 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 3 |
Hb_001195_400 |
0.0702886402 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 4 |
Hb_001818_100 |
0.0712849007 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 5 |
Hb_005976_080 |
0.0778280381 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 6 |
Hb_000185_220 |
0.0795715518 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 7 |
Hb_003305_040 |
0.0804240679 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 8 |
Hb_012393_030 |
0.0813114467 |
- |
- |
NADP-dependent isocitrate dehydrogenase family protein [Populus trichocarpa] |
| 9 |
Hb_002392_010 |
0.0822643081 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 10 |
Hb_007533_040 |
0.0836182795 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_000080_130 |
0.0857486802 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 12 |
Hb_004108_160 |
0.0869506591 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 13 |
Hb_170138_010 |
0.0889046217 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003025_100 |
0.0900713249 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006059_010 |
0.0916480524 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 16 |
Hb_000841_050 |
0.0916793656 |
- |
- |
hypothetical protein L484_019972 [Morus notabilis] |
| 17 |
Hb_004159_020 |
0.0916874692 |
- |
- |
Putative dihydrodipicolinate reductase 2, chloroplastic [Aegilops tauschii] |
| 18 |
Hb_006059_030 |
0.0917622483 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_001541_120 |
0.0920148636 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 20 |
Hb_002748_020 |
0.0922984559 |
- |
- |
transducin family protein [Populus trichocarpa] |