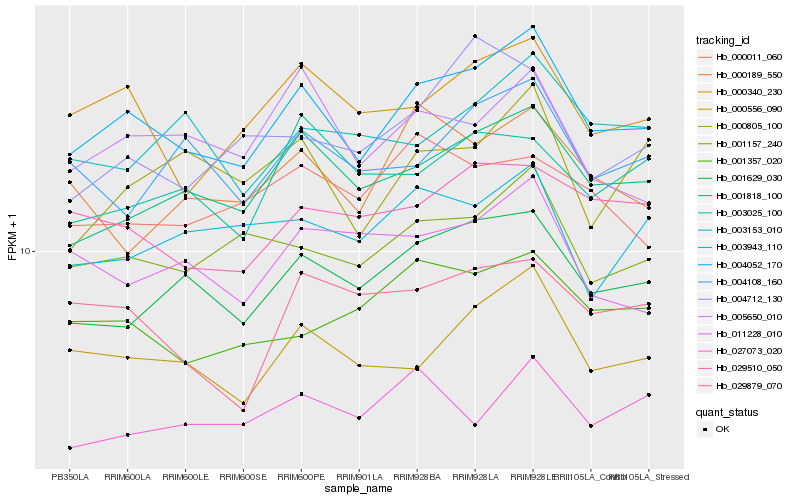
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001818_100 |
0.0665701117 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 3 |
Hb_000556_090 |
0.0685622468 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001629_030 |
0.0709783456 |
- |
- |
PAP-specific phosphatase HAL2-like family protein [Populus trichocarpa] |
| 5 |
Hb_001157_240 |
0.0739738111 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 6 |
Hb_000340_230 |
0.0753048149 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 7 |
Hb_029510_050 |
0.0847685135 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 8 |
Hb_001357_020 |
0.0862359512 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005650_010 |
0.0873426688 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_004108_160 |
0.0908935454 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle-like [Jatropha curcas] |
| 11 |
Hb_000805_100 |
0.0918931949 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_003153_010 |
0.0926243848 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 13 |
Hb_003943_110 |
0.0936646645 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 14 |
Hb_000189_550 |
0.0939079831 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 15 |
Hb_011228_010 |
0.0945493031 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_003025_100 |
0.0946862277 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_029879_070 |
0.0947714823 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 18 |
Hb_027073_020 |
0.0954325181 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 19 |
Hb_000011_060 |
0.0956438567 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004712_130 |
0.0959848107 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |