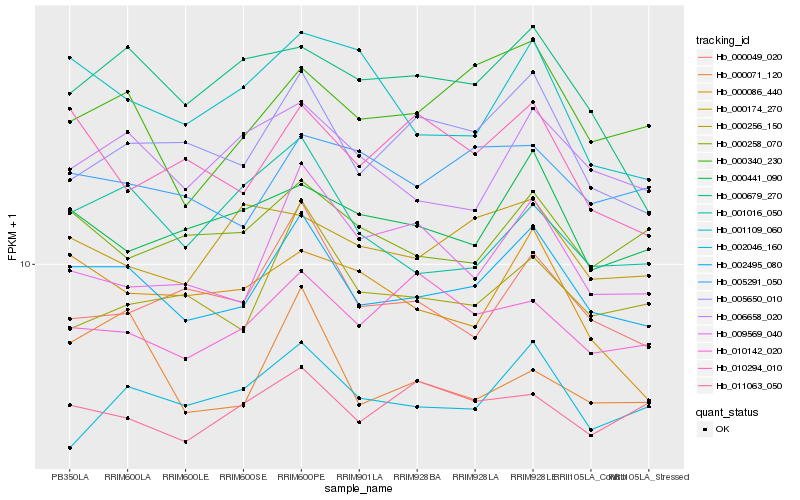
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002495_080 |
0.0 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 2 |
Hb_005650_010 |
0.0748577922 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_006658_020 |
0.0774979722 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 4 |
Hb_000340_230 |
0.0781458831 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 5 |
Hb_000441_090 |
0.0808253899 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 6 |
Hb_000258_070 |
0.081806084 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 7 |
Hb_000049_020 |
0.0821711751 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000256_150 |
0.0827278248 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 9 |
Hb_005291_050 |
0.0830771114 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_009569_040 |
0.0858962695 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 11 |
Hb_002046_160 |
0.0867759629 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_010142_020 |
0.0888085406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011063_050 |
0.0894080283 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 14 |
Hb_001016_050 |
0.0894916646 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 15 |
Hb_000071_120 |
0.0903998771 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 16 |
Hb_000679_270 |
0.0904646953 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 17 |
Hb_010294_010 |
0.0906808239 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 18 |
Hb_001109_060 |
0.0908787302 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_000174_270 |
0.091433197 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000086_440 |
0.0919341134 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |