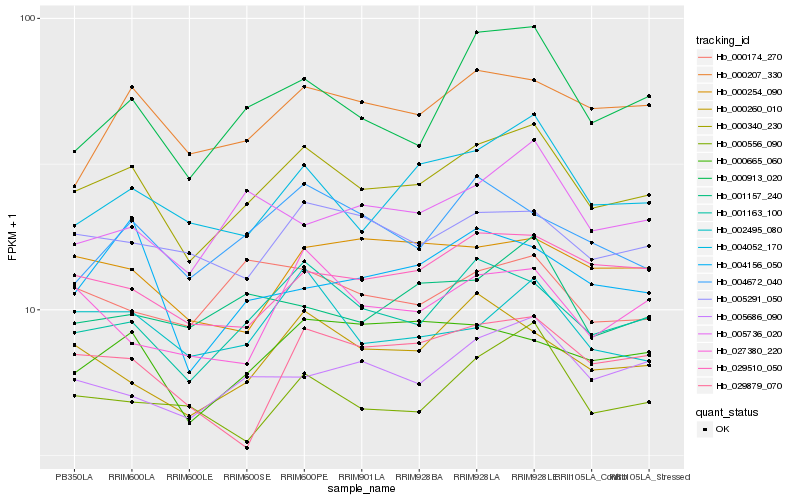
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_230 |
0.0 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 2 |
Hb_001163_100 |
0.0591778121 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_004052_170 |
0.0753048149 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000913_020 |
0.077070248 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 5 |
Hb_029510_050 |
0.0780494134 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 6 |
Hb_002495_080 |
0.0781458831 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 7 |
Hb_005686_090 |
0.0786812452 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_010 |
0.0801345609 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 9 |
Hb_005291_050 |
0.0817808962 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000665_060 |
0.0821191393 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 11 |
Hb_000556_090 |
0.0822399274 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000207_330 |
0.0843125284 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 13 |
Hb_005736_020 |
0.0855290905 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 14 |
Hb_029879_070 |
0.085614373 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 15 |
Hb_001157_240 |
0.0857830796 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 16 |
Hb_004672_040 |
0.0867671591 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 17 |
Hb_000174_270 |
0.0885523697 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_027380_220 |
0.0886022664 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 19 |
Hb_000254_090 |
0.0886820564 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |
| 20 |
Hb_004156_050 |
0.0893186265 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |