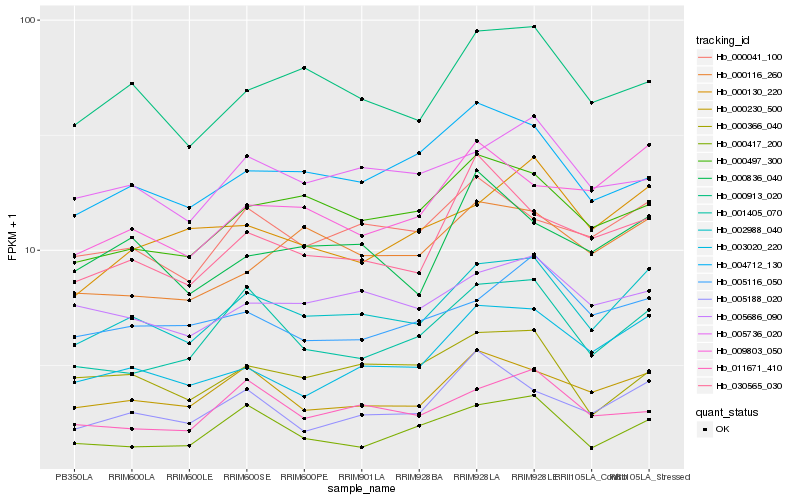
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002988_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000230_500 |
0.0787381688 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000913_020 |
0.081033847 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000497_300 |
0.0845201642 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 5 |
Hb_003020_220 |
0.0853217551 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000041_100 |
0.0858700171 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 7 |
Hb_005686_090 |
0.0876224065 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001405_070 |
0.0895287553 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 9 |
Hb_005736_020 |
0.0906522166 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 10 |
Hb_005116_050 |
0.0933765231 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000130_220 |
0.0938485975 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000116_260 |
0.0939704972 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 13 |
Hb_009803_050 |
0.0975041954 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 14 |
Hb_005188_020 |
0.0976162443 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 15 |
Hb_004712_130 |
0.0984713718 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 16 |
Hb_000366_040 |
0.0996249829 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 17 |
Hb_000836_040 |
0.101188037 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_011671_410 |
0.1017470963 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 19 |
Hb_030565_030 |
0.1027671323 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 20 |
Hb_000417_200 |
0.1032249207 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |