| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
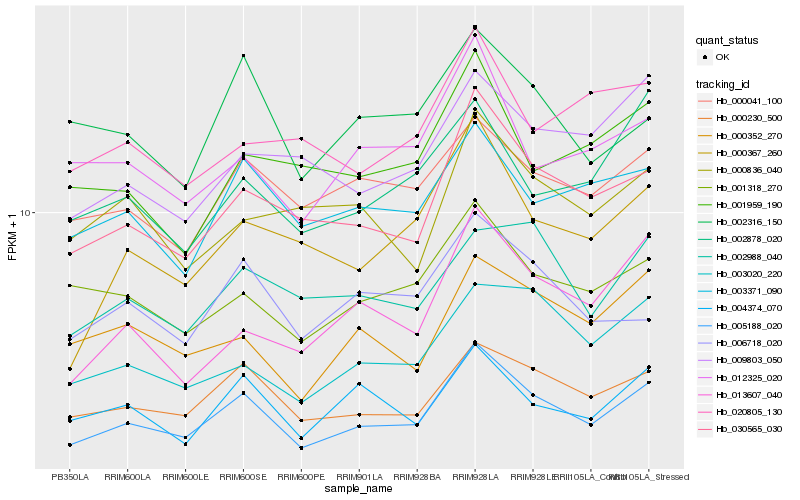
Hb_005188_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 2 |
Hb_000230_500 |
0.0655908362 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 3 |
Hb_030565_030 |
0.066921188 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 4 |
Hb_000041_100 |
0.086125132 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 5 |
Hb_004374_070 |
0.0869212881 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 6 |
Hb_001318_270 |
0.0871753285 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_003371_090 |
0.088415691 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 8 |
Hb_000352_270 |
0.0934029069 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_012325_020 |
0.0949844984 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003020_220 |
0.095024087 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006718_020 |
0.0965626999 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002988_040 |
0.0976162443 |
- |
- |
- |
| 13 |
Hb_009803_050 |
0.0988459449 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 14 |
Hb_000367_260 |
0.0997241999 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 15 |
Hb_002316_150 |
0.1000910193 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 16 |
Hb_013607_040 |
0.1002410565 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 17 |
Hb_002878_020 |
0.1003493202 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 18 |
Hb_001959_190 |
0.1025937128 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 19 |
Hb_000836_040 |
0.1040226139 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_020805_130 |
0.1069480046 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |