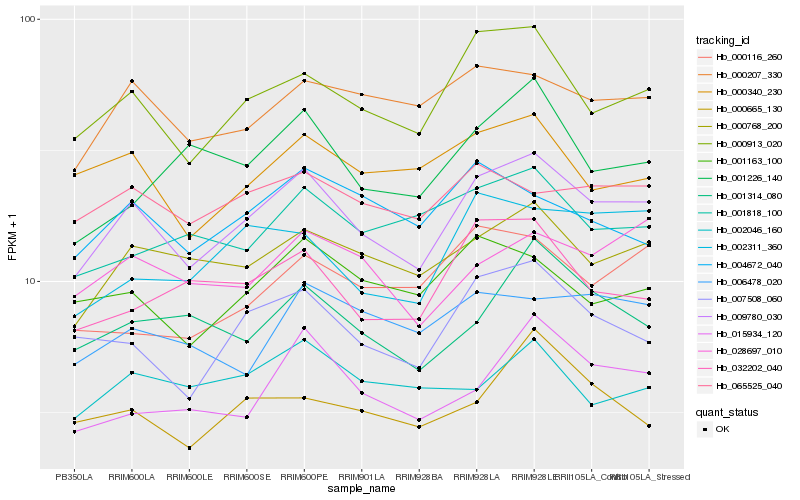
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009780_030 |
0.0 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 2 |
Hb_000913_020 |
0.0673983825 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000768_200 |
0.0817746747 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 4 |
Hb_007508_060 |
0.0846383201 |
- |
- |
PREDICTED: uncharacterized protein LOC105640628 [Jatropha curcas] |
| 5 |
Hb_000207_330 |
0.0911088644 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 6 |
Hb_028697_010 |
0.0984036127 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 7 |
Hb_001163_100 |
0.098651614 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000340_230 |
0.0988049005 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 9 |
Hb_002311_360 |
0.0998452706 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 10 |
Hb_032202_040 |
0.100404506 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 11 |
Hb_065525_040 |
0.1013107473 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 12 |
Hb_001314_080 |
0.1029363509 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 13 |
Hb_015934_120 |
0.1034394983 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 14 |
Hb_001818_100 |
0.1044866212 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 15 |
Hb_004672_040 |
0.1054190834 |
- |
- |
PREDICTED: SEC1 family transport protein SLY1-like [Populus euphratica] |
| 16 |
Hb_006478_020 |
0.1054556321 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_001226_140 |
0.1094140887 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 18 |
Hb_000116_260 |
0.1100149812 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 19 |
Hb_002046_160 |
0.1101136002 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000665_130 |
0.1106756049 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |