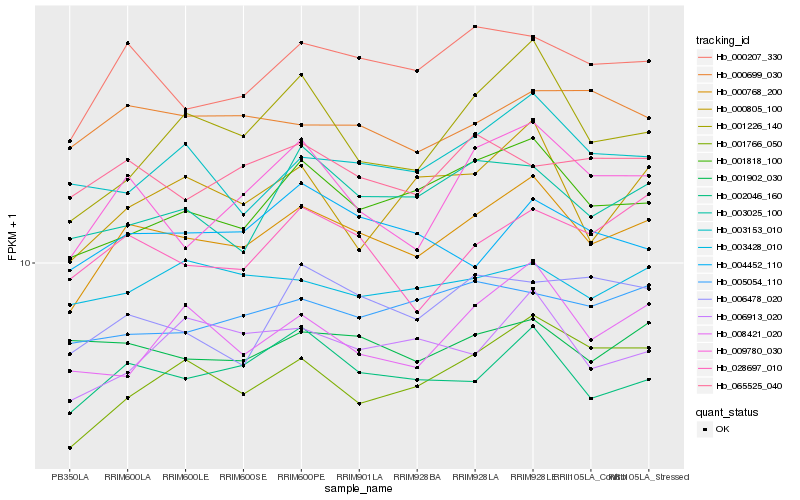
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000768_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 2 |
Hb_000207_330 |
0.0674270262 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 3 |
Hb_002046_160 |
0.0680319926 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001818_100 |
0.0728110325 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 5 |
Hb_009780_030 |
0.0817746747 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 6 |
Hb_001766_050 |
0.0849269619 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 7 |
Hb_003025_100 |
0.0865793185 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006913_020 |
0.0876742676 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 9 |
Hb_028697_010 |
0.088254075 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 10 |
Hb_003153_010 |
0.0892692243 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like [Jatropha curcas] |
| 11 |
Hb_000805_100 |
0.0899959231 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_000699_030 |
0.0913158593 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_005054_110 |
0.0913428133 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 14 |
Hb_001226_140 |
0.0924044301 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 15 |
Hb_003428_010 |
0.0927100677 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 16 |
Hb_008421_020 |
0.0933262629 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001902_030 |
0.0933640539 |
- |
- |
PREDICTED: SKP1-like protein 21 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004452_110 |
0.0934337997 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 19 |
Hb_065525_040 |
0.0936899621 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 20 |
Hb_006478_020 |
0.0943853806 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |