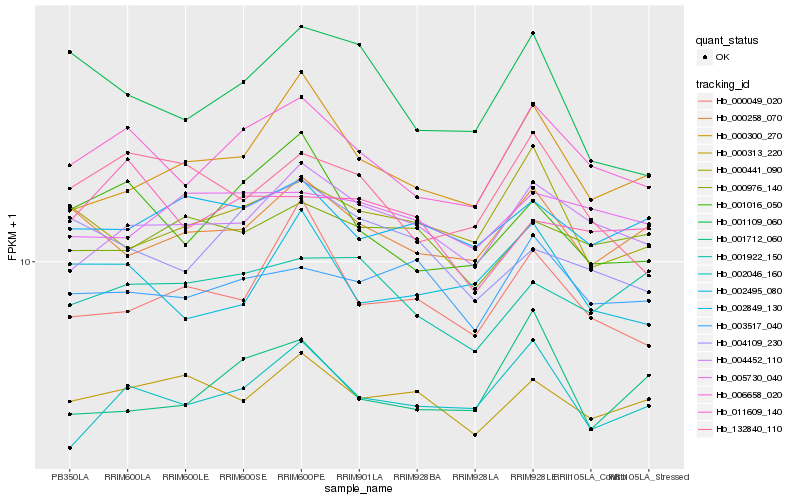
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006658_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 2 |
Hb_001016_050 |
0.0611039243 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 3 |
Hb_004452_110 |
0.0654746229 |
- |
- |
PREDICTED: uncharacterized protein LOC105639575 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002046_160 |
0.0717741434 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000258_070 |
0.0773014418 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 6 |
Hb_002495_080 |
0.0774979722 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 7 |
Hb_132840_110 |
0.0786142279 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_000300_270 |
0.0802109503 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_003517_040 |
0.0807058366 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000313_220 |
0.081532307 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000441_090 |
0.0819007856 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 12 |
Hb_001922_150 |
0.0829448589 |
- |
- |
PREDICTED: WD repeat-containing protein 91 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_011609_140 |
0.0842902607 |
- |
- |
hypothetical protein JCGZ_12974 [Jatropha curcas] |
| 14 |
Hb_004109_230 |
0.0843008711 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_002849_130 |
0.0857304554 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 16 |
Hb_000049_020 |
0.0870829833 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 17 |
Hb_005730_040 |
0.0876448023 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 18 |
Hb_001712_060 |
0.0886600915 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 19 |
Hb_000976_140 |
0.0888907791 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001109_060 |
0.0889838548 |
- |
- |
amino acid transporter, putative [Ricinus communis] |