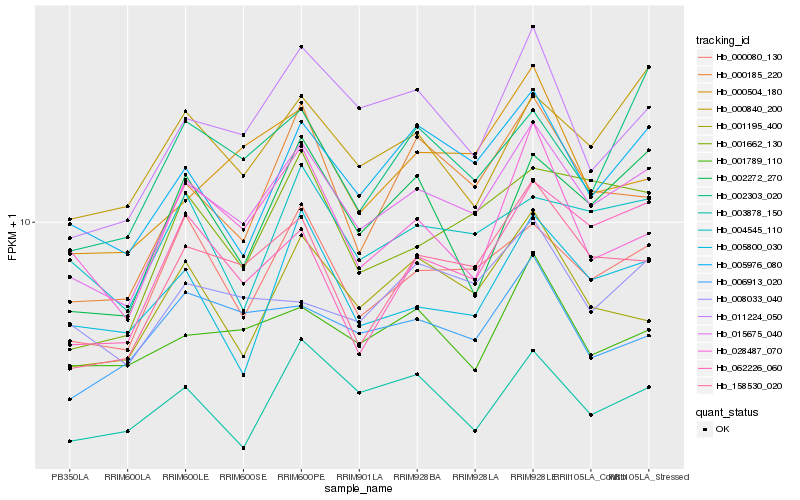
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015675_040 |
0.0 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 2 |
Hb_000080_130 |
0.0614545183 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 3 |
Hb_158530_020 |
0.0616252854 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 4 |
Hb_005976_080 |
0.0672462509 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 5 |
Hb_011224_050 |
0.0678114083 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 6 |
Hb_000840_200 |
0.0749245198 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000185_220 |
0.076862595 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 8 |
Hb_001195_400 |
0.0771019658 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 9 |
Hb_003878_150 |
0.0784537503 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 10 |
Hb_001662_130 |
0.0786583381 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002272_270 |
0.0799291369 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_004545_110 |
0.0799766539 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_062226_060 |
0.0804907344 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 14 |
Hb_008033_040 |
0.0806099163 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000504_180 |
0.0817631142 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 16 |
Hb_028487_070 |
0.0821654292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001789_110 |
0.0828434503 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_002303_020 |
0.0828990499 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 19 |
Hb_005800_030 |
0.083468669 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 20 |
Hb_006913_020 |
0.0843267249 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |