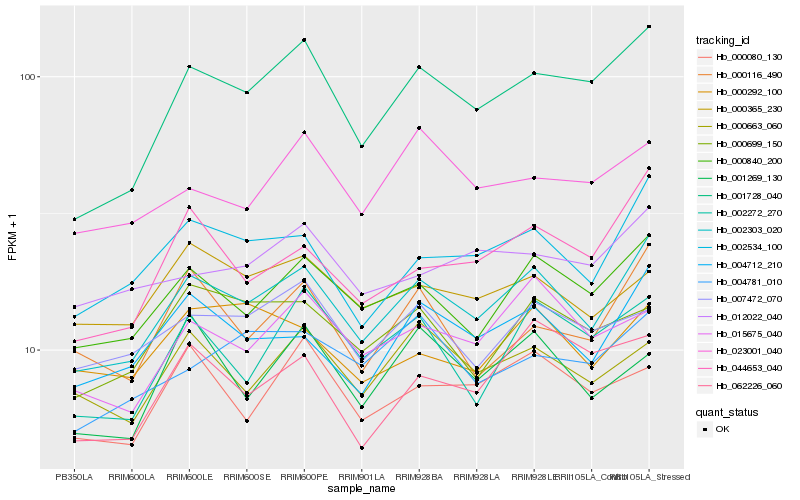
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002303_020 |
0.0 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Gossypium raimondii] |
| 2 |
Hb_000840_200 |
0.0595315932 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_002534_100 |
0.0647743273 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004712_210 |
0.0647927664 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM50-like [Jatropha curcas] |
| 5 |
Hb_001728_040 |
0.0679018082 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 6 |
Hb_044653_040 |
0.078861963 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 7 |
Hb_015675_040 |
0.0828990499 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 8 |
Hb_000663_060 |
0.085223598 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 9 |
Hb_000116_490 |
0.0862028501 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 10 |
Hb_000699_150 |
0.0887082145 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 11 |
Hb_002272_270 |
0.0887167626 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_007472_070 |
0.0894160068 |
- |
- |
cir, putative [Ricinus communis] |
| 13 |
Hb_000080_130 |
0.0901964753 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 14 |
Hb_062226_060 |
0.091643996 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 15 |
Hb_004781_010 |
0.0919581551 |
- |
- |
PREDICTED: potassium transporter 11-like [Populus euphratica] |
| 16 |
Hb_000292_100 |
0.0923540585 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 17 |
Hb_000365_230 |
0.0923727011 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 18 |
Hb_012022_040 |
0.0931058392 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 19 |
Hb_001269_130 |
0.0934165329 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 20 |
Hb_023001_040 |
0.0937061362 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |