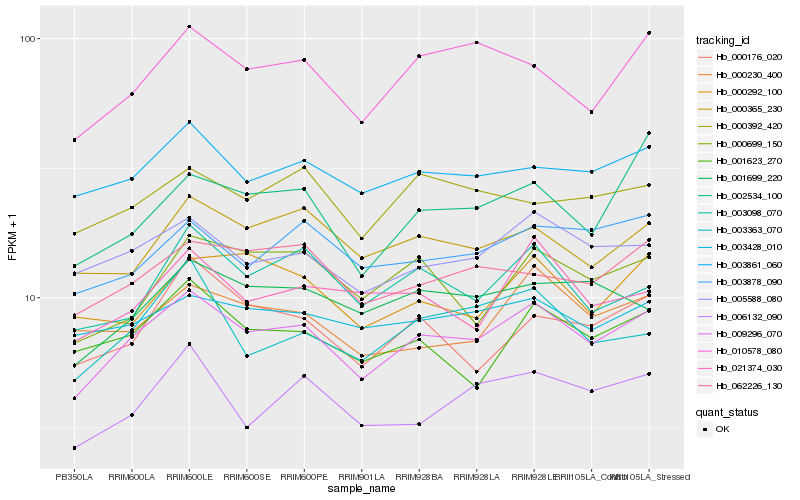
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009296_070 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005588_080 |
0.0680874189 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 3 |
Hb_003861_060 |
0.0709104868 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |
| 4 |
Hb_010578_080 |
0.0725025814 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 5 |
Hb_062226_130 |
0.0738216338 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 6 |
Hb_003878_090 |
0.0773409541 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 7 |
Hb_000365_230 |
0.0774631257 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 8 |
Hb_001699_220 |
0.0786790842 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 9 |
Hb_003428_010 |
0.0798766197 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 10 |
Hb_006132_090 |
0.0799850936 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003098_070 |
0.0805641238 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 12 |
Hb_001623_270 |
0.0818120748 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 13 |
Hb_000176_020 |
0.0824765433 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 14 |
Hb_000230_400 |
0.0830516979 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003363_070 |
0.0831616338 |
- |
- |
- |
| 16 |
Hb_000699_150 |
0.0831893298 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 17 |
Hb_000392_420 |
0.0836148244 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 18 |
Hb_002534_100 |
0.0846124955 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000292_100 |
0.0846951999 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 20 |
Hb_021374_030 |
0.0852020497 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |