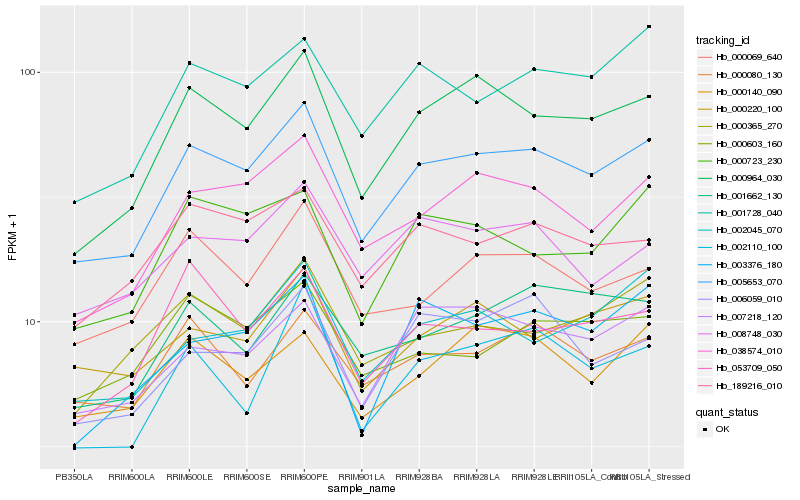
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005653_070 |
0.0 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 2 |
Hb_000964_030 |
0.0556267547 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 3 |
Hb_038574_010 |
0.0604794303 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like isoform X2 [Populus euphratica] |
| 4 |
Hb_189216_010 |
0.0755098141 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001728_040 |
0.0758790521 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 6 |
Hb_007218_120 |
0.07788168 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g51965, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000069_640 |
0.0788718447 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 8 |
Hb_002110_100 |
0.080217248 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000080_130 |
0.0802480451 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 10 |
Hb_000723_230 |
0.0813025128 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 11 |
Hb_000220_100 |
0.0823496637 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 [Jatropha curcas] |
| 12 |
Hb_001662_130 |
0.0824984684 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_008748_030 |
0.0836466712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006059_010 |
0.0841126834 |
- |
- |
PREDICTED: uncharacterized protein LOC105648671 [Jatropha curcas] |
| 15 |
Hb_000140_090 |
0.0843219297 |
- |
- |
PREDICTED: C-terminal binding protein AN [Jatropha curcas] |
| 16 |
Hb_000365_270 |
0.0843237201 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 17 |
Hb_002045_070 |
0.0847555684 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 18 |
Hb_000603_160 |
0.0848346899 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 19 |
Hb_053709_050 |
0.0852896748 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_003376_180 |
0.0854877515 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |