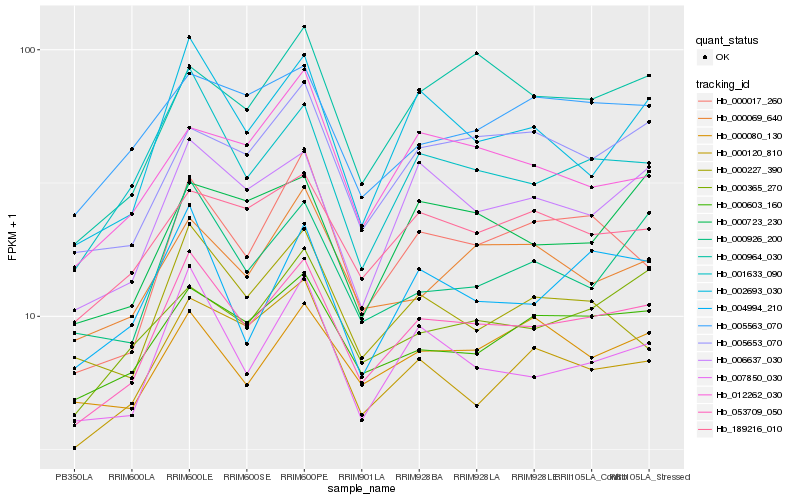
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053709_050 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_001633_090 |
0.0695282191 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 3 |
Hb_007850_030 |
0.0704554567 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 4 |
Hb_006637_030 |
0.0743339508 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 5 |
Hb_000603_160 |
0.0756350399 |
- |
- |
PREDICTED: fatty-acid-binding protein 2 [Jatropha curcas] |
| 6 |
Hb_002693_030 |
0.0799097924 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 7 |
Hb_189216_010 |
0.0800178849 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000926_200 |
0.0819970293 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_004994_210 |
0.0832475071 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 10 |
Hb_000365_270 |
0.08519256 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 11 |
Hb_005653_070 |
0.0852896748 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 12 |
Hb_005563_070 |
0.0857014812 |
- |
- |
PREDICTED: uncharacterized protein LOC105630108 [Jatropha curcas] |
| 13 |
Hb_000069_640 |
0.0872004728 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 14 |
Hb_000017_260 |
0.0872785313 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 15 |
Hb_000120_810 |
0.0893361257 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 16 |
Hb_000964_030 |
0.0895658325 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 17 |
Hb_000723_230 |
0.0944275931 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 18 |
Hb_000227_390 |
0.0957509937 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000080_130 |
0.0964030794 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 20 |
Hb_012262_030 |
0.0978646345 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |