| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
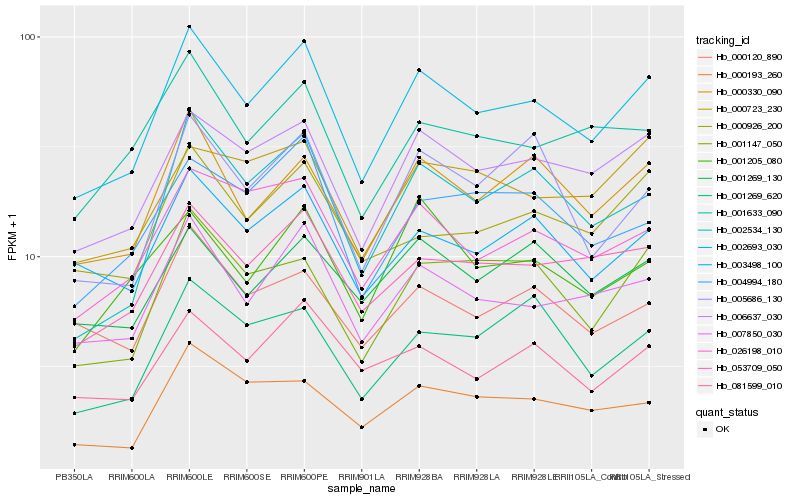
Hb_002693_030 |
0.0 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 2 |
Hb_006637_030 |
0.0660277639 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 3 |
Hb_007850_030 |
0.0676875591 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 4 |
Hb_000330_090 |
0.0795121116 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 5 |
Hb_053709_050 |
0.0799097924 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_002534_130 |
0.0827480966 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 7 |
Hb_026198_010 |
0.0849745124 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003498_100 |
0.0902414465 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 9 |
Hb_081599_010 |
0.0929014347 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001147_050 |
0.093300604 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000926_200 |
0.0969732904 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001633_090 |
0.0977922419 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 13 |
Hb_004994_180 |
0.0995823685 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 14 |
Hb_000193_260 |
0.0997430451 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 15 |
Hb_000120_890 |
0.1014756291 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 16 |
Hb_005686_130 |
0.1022163017 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 17 |
Hb_001269_130 |
0.1026093889 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 18 |
Hb_001205_080 |
0.1036735566 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |
| 19 |
Hb_000723_230 |
0.1037687433 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 20 |
Hb_001269_620 |
0.1048887632 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |