| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
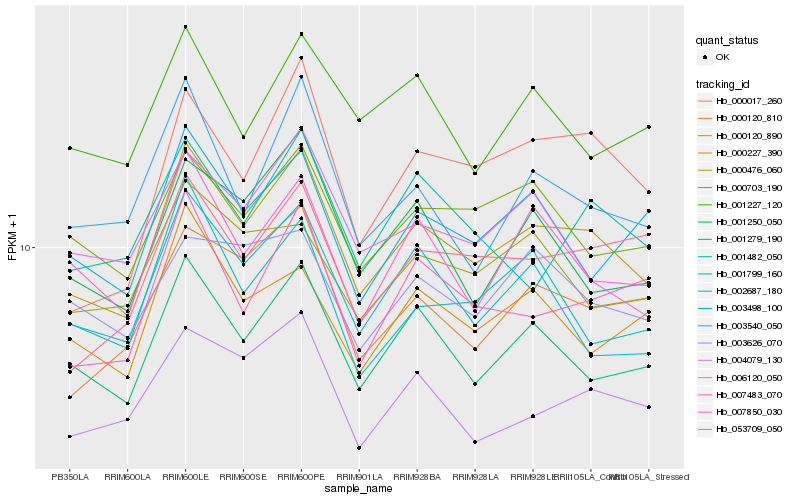
Hb_000227_390 |
0.0 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001279_190 |
0.0716168196 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007483_070 |
0.0845578084 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 4 |
Hb_000120_810 |
0.0872560606 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 5 |
Hb_004079_130 |
0.0879893064 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 6 |
Hb_003540_050 |
0.088222174 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 7 |
Hb_000703_190 |
0.0898413394 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007850_030 |
0.0903901947 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 9 |
Hb_002687_180 |
0.0904604898 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 10 |
Hb_001250_050 |
0.0911980213 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000017_260 |
0.0922679547 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 12 |
Hb_003498_100 |
0.0942088677 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 13 |
Hb_053709_050 |
0.0957509937 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000476_060 |
0.0959438333 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_006120_050 |
0.096950632 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 16 |
Hb_000120_890 |
0.0971659022 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 17 |
Hb_001799_160 |
0.0979908602 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_001482_050 |
0.0992557947 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 19 |
Hb_001227_120 |
0.0999679598 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 20 |
Hb_003626_070 |
0.1004567333 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |