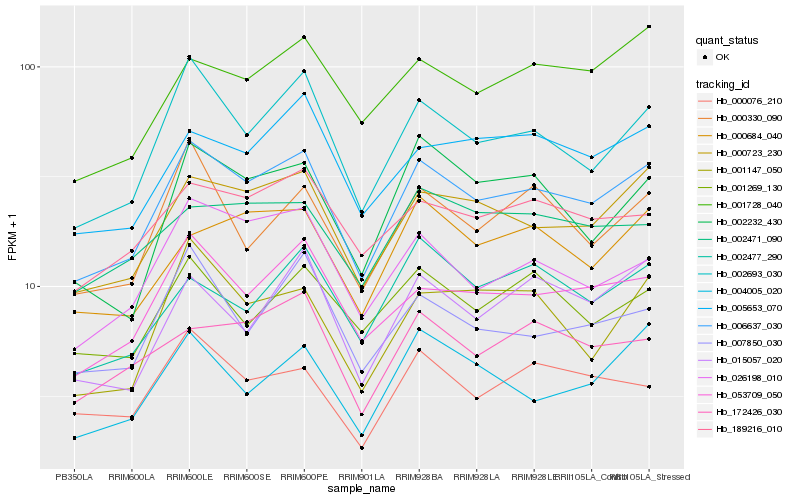
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006637_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 2 |
Hb_000723_230 |
0.0600409311 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 3 |
Hb_002693_030 |
0.0660277639 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 4 |
Hb_053709_050 |
0.0743339508 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_026198_010 |
0.083817667 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 6 |
Hb_189216_010 |
0.0864929617 |
- |
- |
PREDICTED: uncharacterized protein LOC105642236 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005653_070 |
0.0874334349 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 8 |
Hb_001728_040 |
0.089432167 |
- |
- |
Aconitase/3-isopropylmalate dehydratase protein [Theobroma cacao] |
| 9 |
Hb_000684_040 |
0.0908209073 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR1 [Jatropha curcas] |
| 10 |
Hb_002232_430 |
0.0925289625 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000330_090 |
0.0938170137 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 12 |
Hb_172426_030 |
0.0940523757 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001269_130 |
0.0952430537 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 14 |
Hb_000076_210 |
0.0963686034 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_002471_090 |
0.0963891754 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 16 |
Hb_015057_020 |
0.0969214344 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 17 |
Hb_007850_030 |
0.0978972665 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 18 |
Hb_001147_050 |
0.0992515167 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002477_290 |
0.0994800171 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_004005_020 |
0.1011257429 |
- |
- |
PREDICTED: nudix hydrolase 20, chloroplastic-like isoform X1 [Jatropha curcas] |