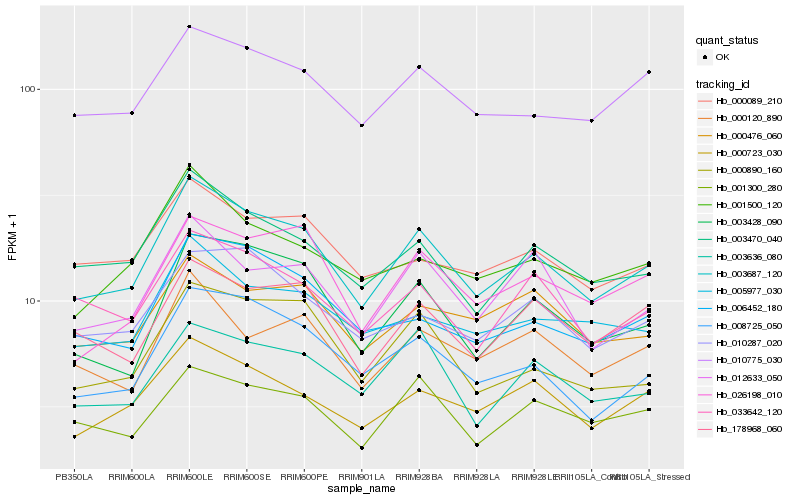
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003687_120 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 2 |
Hb_003470_040 |
0.0705601221 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_008725_050 |
0.0706607238 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_003428_090 |
0.0717798272 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 5 |
Hb_000723_030 |
0.075942389 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X1 [Jatropha curcas] |
| 6 |
Hb_000890_160 |
0.0795019869 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 7 |
Hb_178968_060 |
0.0803774561 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 8 |
Hb_012633_050 |
0.0807961047 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_033642_120 |
0.0808501739 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 10 |
Hb_010775_030 |
0.0818427945 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 11 |
Hb_001300_280 |
0.0822464 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 12 |
Hb_026198_010 |
0.0822922623 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006452_180 |
0.0827585785 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 14 |
Hb_010287_020 |
0.083982809 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 15 |
Hb_005977_030 |
0.0839967386 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 16 |
Hb_003636_080 |
0.0840710753 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 17 |
Hb_000120_890 |
0.0841311102 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 18 |
Hb_001500_120 |
0.0860486144 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_000089_210 |
0.086186192 |
- |
- |
unknown [Medicago truncatula] |
| 20 |
Hb_000476_060 |
0.0862811558 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |