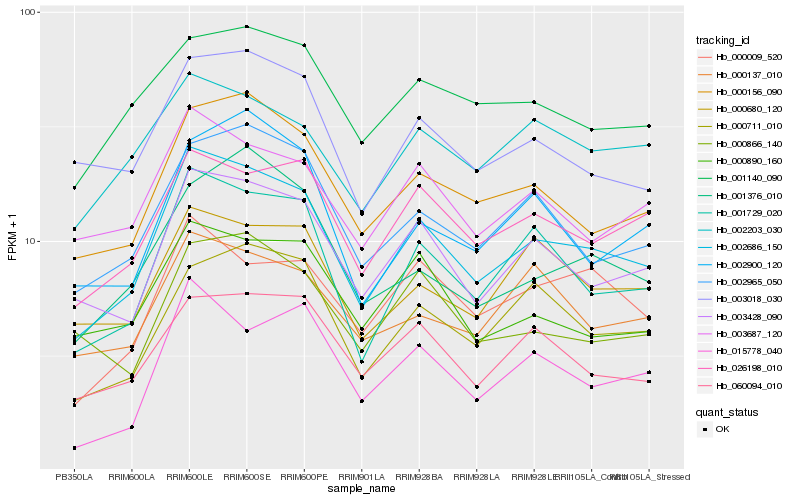
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_150 |
0.0 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 2 |
Hb_001729_020 |
0.0734803328 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_003428_090 |
0.0766428635 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 4 |
Hb_000156_090 |
0.0836682007 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002965_050 |
0.0912958685 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 6 |
Hb_026198_010 |
0.0927659292 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 7 |
Hb_060094_010 |
0.097877948 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003687_120 |
0.0982101114 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 9 |
Hb_003018_030 |
0.1008301053 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 10 |
Hb_000711_010 |
0.1010915403 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 11 |
Hb_000890_160 |
0.1044773174 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 12 |
Hb_000009_520 |
0.1046966373 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 13 |
Hb_000680_120 |
0.1048437355 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 14 |
Hb_000137_010 |
0.1082120441 |
- |
- |
PREDICTED: uncharacterized protein LOC105644349 [Jatropha curcas] |
| 15 |
Hb_001140_090 |
0.1091125737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_015778_040 |
0.1106954553 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 17 |
Hb_002900_120 |
0.1109595161 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 18 |
Hb_001376_010 |
0.1122273378 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 19 |
Hb_002203_030 |
0.112503751 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 20 |
Hb_000866_140 |
0.1130426347 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |