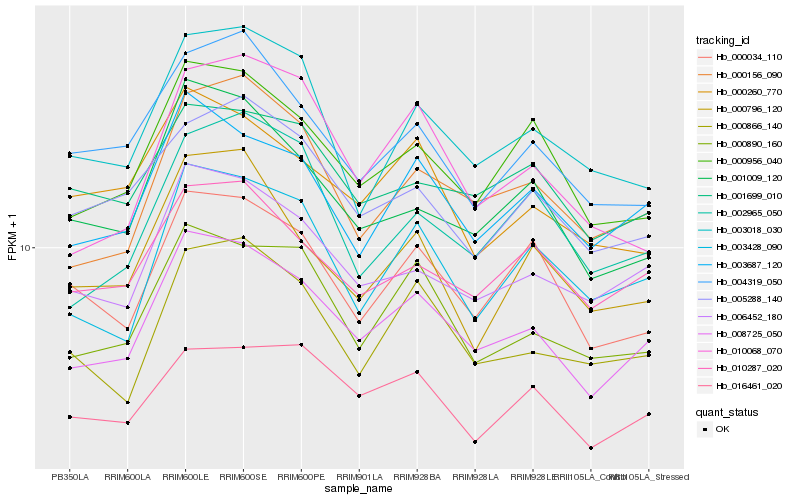
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_050 |
0.0 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 2 |
Hb_003687_120 |
0.0706607238 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 3 |
Hb_006452_180 |
0.0731324978 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 4 |
Hb_000956_040 |
0.0746028469 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 5 |
Hb_000890_160 |
0.0753017991 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 6 |
Hb_000156_090 |
0.0756936534 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003428_090 |
0.0797954184 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 8 |
Hb_001009_120 |
0.0850881965 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 9 |
Hb_010287_020 |
0.0855762367 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 10 |
Hb_000866_140 |
0.0882753186 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 11 |
Hb_000034_110 |
0.0889257138 |
- |
- |
sentrin/sumo-specific protease, putative [Ricinus communis] |
| 12 |
Hb_016461_020 |
0.0908011801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001699_010 |
0.0935515556 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 14 |
Hb_003018_030 |
0.0937279067 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 15 |
Hb_005288_140 |
0.0949586307 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 16 |
Hb_004319_050 |
0.0953365602 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 17 |
Hb_000260_770 |
0.0974668493 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 18 |
Hb_002965_050 |
0.0975607091 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 19 |
Hb_000796_120 |
0.0982146083 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_010068_070 |
0.0988675178 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |