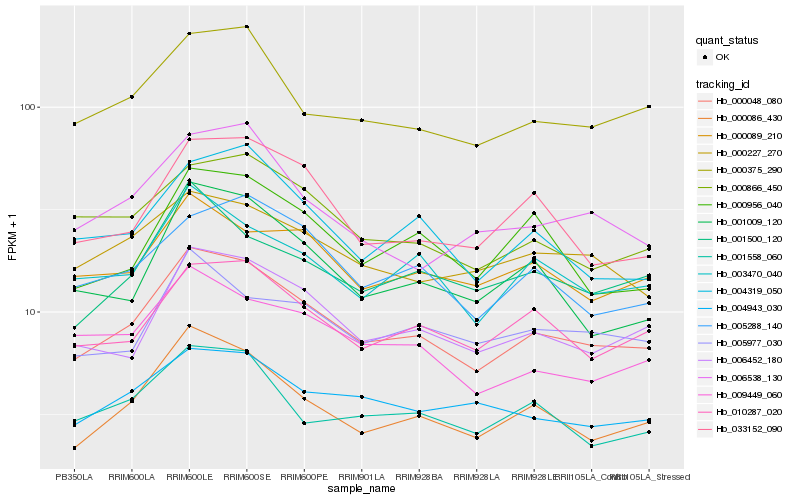
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000048_080 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000086_430 |
0.0663375504 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 3 |
Hb_006452_180 |
0.0686896657 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 4 |
Hb_000375_290 |
0.0755618054 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_010287_020 |
0.0766058357 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 6 |
Hb_003470_040 |
0.0791547865 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_004319_050 |
0.0831912759 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 8 |
Hb_005288_140 |
0.0833824253 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta-like [Jatropha curcas] |
| 9 |
Hb_033152_090 |
0.0848967835 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2E isoform X2 [Gossypium raimondii] |
| 10 |
Hb_000866_450 |
0.0850613857 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_001009_120 |
0.0870362252 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001500_120 |
0.0874820576 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_009449_060 |
0.0878451124 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 14 |
Hb_004943_030 |
0.0883648063 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 15 |
Hb_000227_270 |
0.0884359548 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001558_060 |
0.0893724734 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000956_040 |
0.0898772766 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 18 |
Hb_000089_210 |
0.0903136396 |
- |
- |
unknown [Medicago truncatula] |
| 19 |
Hb_005977_030 |
0.091501216 |
- |
- |
PREDICTED: monothiol glutaredoxin-S10-like [Jatropha curcas] |
| 20 |
Hb_006538_130 |
0.0918492311 |
- |
- |
Uncharacterized protein TCM_009867 [Theobroma cacao] |