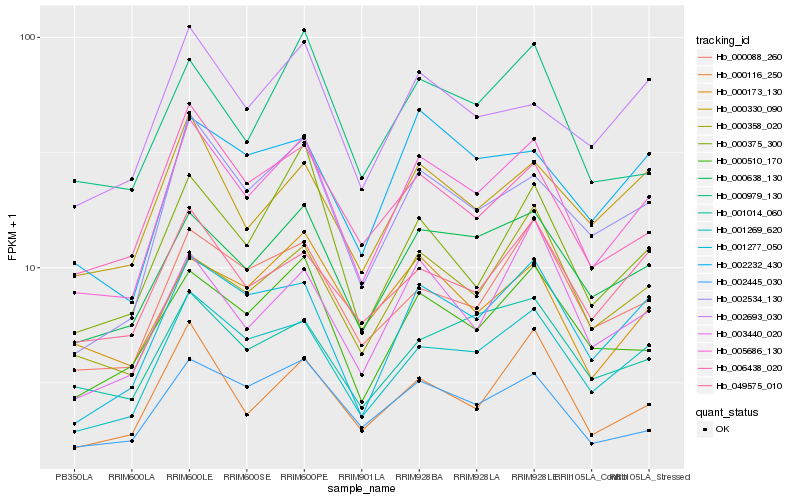
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005686_130 |
0.0 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 2 |
Hb_001269_620 |
0.0694731256 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_000638_130 |
0.0753819113 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 4 |
Hb_001277_050 |
0.0815866376 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002534_130 |
0.0827696416 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 6 |
Hb_000510_170 |
0.0884478001 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 7 |
Hb_000088_260 |
0.0915649364 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 8 |
Hb_002445_030 |
0.0929678067 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 9 |
Hb_000116_250 |
0.0941667687 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 10 |
Hb_006438_020 |
0.0964352579 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 11 |
Hb_000330_090 |
0.0987736179 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000358_020 |
0.1004157277 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 13 |
Hb_002232_430 |
0.1014044982 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002693_030 |
0.1022163017 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 15 |
Hb_003440_020 |
0.1032656663 |
- |
- |
PREDICTED: serine protease SPPA, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_049575_010 |
0.1035099986 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 17 |
Hb_000375_300 |
0.1041700591 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 18 |
Hb_000173_130 |
0.1043465218 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000979_130 |
0.1052204999 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 20 |
Hb_001014_060 |
0.1062235502 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |