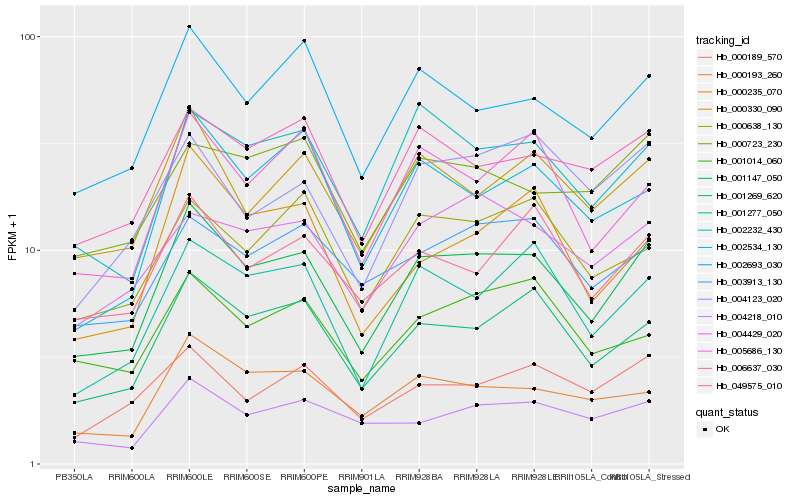
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001147_050 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001269_620 |
0.0797369399 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_000330_090 |
0.0918312053 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002693_030 |
0.093300604 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 5 |
Hb_002232_430 |
0.0972353602 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001277_050 |
0.0984387065 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006637_030 |
0.0992515167 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 8 |
Hb_000193_260 |
0.1009699385 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 9 |
Hb_005686_130 |
0.1065105348 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 10 |
Hb_049575_010 |
0.1085091864 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 11 |
Hb_002534_130 |
0.1097957667 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 12 |
Hb_004218_010 |
0.1118899136 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000189_570 |
0.1134880582 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 14 |
Hb_003913_130 |
0.1139982534 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 15 |
Hb_004429_020 |
0.1145142819 |
- |
- |
PREDICTED: magnesium transporter MRS2-5 [Jatropha curcas] |
| 16 |
Hb_000723_230 |
0.1164554384 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 17 |
Hb_000638_130 |
0.1175436878 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 18 |
Hb_000235_070 |
0.1181518884 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 19 |
Hb_001014_060 |
0.1182331964 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 20 |
Hb_004123_020 |
0.119141546 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |