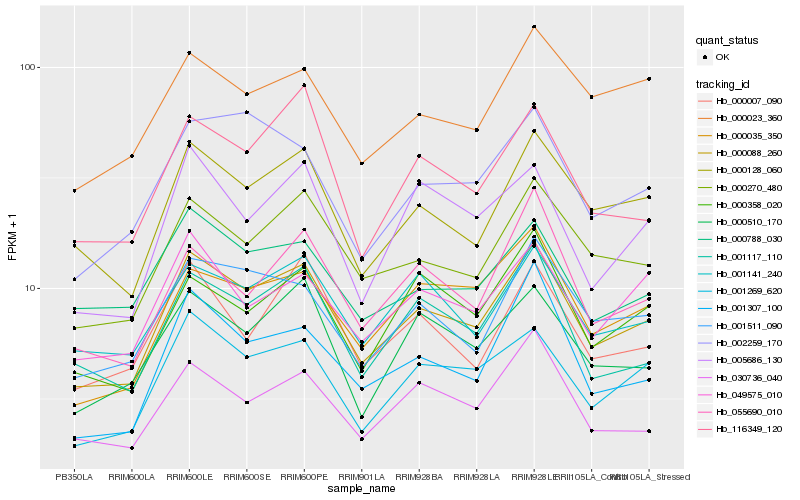
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 2 |
Hb_000270_480 |
0.0685866091 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 3 |
Hb_001269_620 |
0.0697943124 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 4 |
Hb_000128_060 |
0.0717947669 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 5 |
Hb_001511_090 |
0.0767051431 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 6 |
Hb_001117_110 |
0.0785249275 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 7 |
Hb_001141_240 |
0.0810648817 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_030736_040 |
0.0823980648 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000035_350 |
0.0837205651 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001307_100 |
0.0864468013 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_000510_170 |
0.0866401621 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 12 |
Hb_000358_020 |
0.0895748466 |
- |
- |
PREDICTED: uncharacterized protein C05D11.7-like [Jatropha curcas] |
| 13 |
Hb_116349_120 |
0.0897423637 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_000788_030 |
0.0901213847 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 15 |
Hb_055690_010 |
0.0905944772 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_049575_010 |
0.0912334241 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 17 |
Hb_005686_130 |
0.0915649364 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 18 |
Hb_002259_170 |
0.0917369226 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000007_090 |
0.0929196427 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 20 |
Hb_000023_360 |
0.0934848549 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |