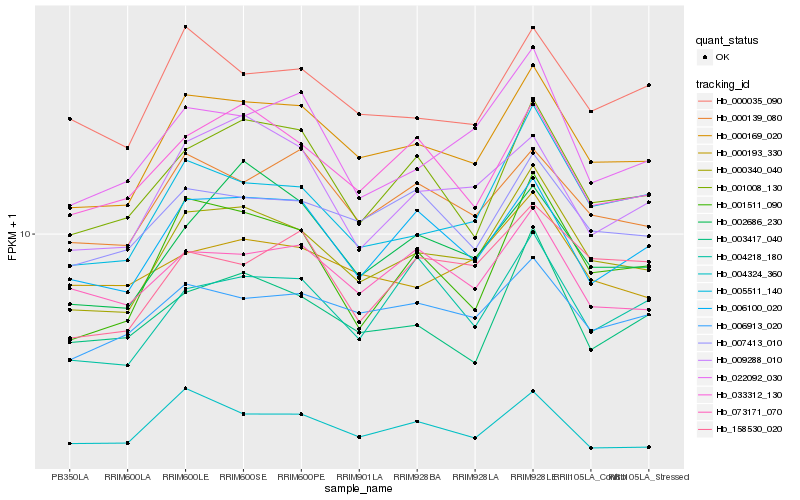
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_040 |
0.0 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000169_020 |
0.040145187 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 3 |
Hb_005511_140 |
0.063478655 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 4 |
Hb_022092_030 |
0.0764722632 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001511_090 |
0.0766804848 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 6 |
Hb_006100_020 |
0.0785760589 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 7 |
Hb_001008_130 |
0.0797572123 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 8 |
Hb_007413_010 |
0.0834255018 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 9 |
Hb_033312_130 |
0.0838702581 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006913_020 |
0.0843281829 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003417_040 |
0.0847618224 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 12 |
Hb_000193_330 |
0.0876809388 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 13 |
Hb_002686_230 |
0.0886263562 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_009288_010 |
0.0893041323 |
desease resistance |
Gene Name: Clp_N |
ERD1 protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000139_080 |
0.0894481089 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000035_090 |
0.0897528603 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 17 |
Hb_158530_020 |
0.0904633699 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 18 |
Hb_004324_360 |
0.0910332814 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 19 |
Hb_073171_070 |
0.0912619393 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_004218_180 |
0.0916805891 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |