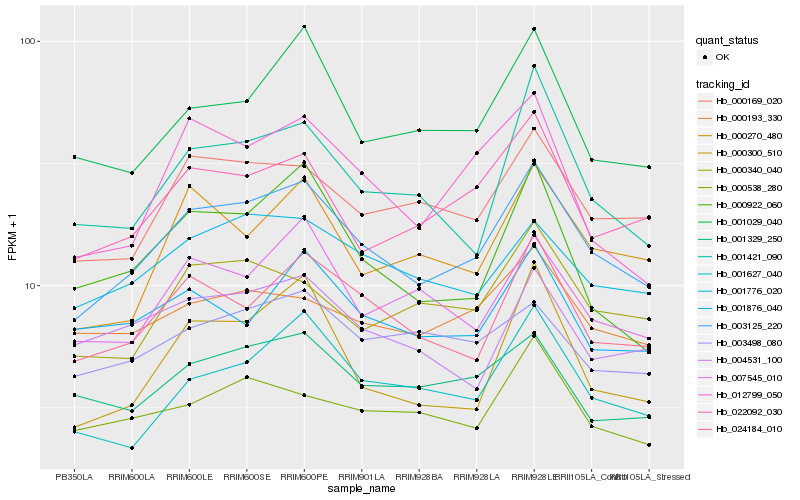
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003125_220 |
0.0 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 2 |
Hb_024184_010 |
0.0790558948 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 3 |
Hb_000169_020 |
0.0818705346 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 4 |
Hb_001776_020 |
0.0864855122 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 5 |
Hb_022092_030 |
0.088734293 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_001627_040 |
0.0931982726 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007545_010 |
0.0942075586 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 8 |
Hb_000340_040 |
0.0957566658 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001029_040 |
0.0959878935 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 10 |
Hb_000270_480 |
0.0961189647 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 11 |
Hb_001421_090 |
0.0980546978 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 12 |
Hb_001329_250 |
0.0992189508 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000193_330 |
0.1018464768 |
- |
- |
PREDICTED: formin-like protein 20 [Jatropha curcas] |
| 14 |
Hb_000538_280 |
0.1022799888 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 15 |
Hb_003498_080 |
0.1031330766 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 16 |
Hb_004531_100 |
0.1034287265 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000922_060 |
0.1037704466 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 18 |
Hb_012799_050 |
0.1039931677 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 19 |
Hb_001876_040 |
0.1043634222 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 20 |
Hb_000300_510 |
0.1047372512 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |