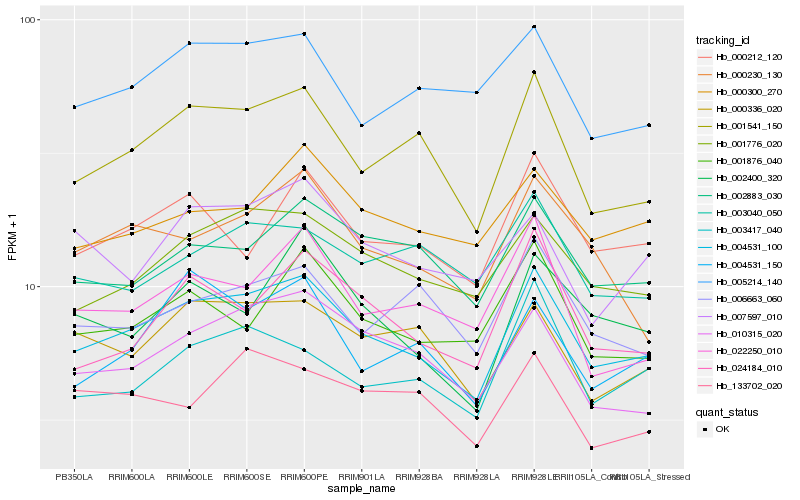
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_100 |
0.0 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001541_150 |
0.0643222024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001776_020 |
0.0721596646 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 4 |
Hb_010315_020 |
0.0800471139 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 5 |
Hb_002883_030 |
0.0835179634 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 6 |
Hb_000336_020 |
0.0854780526 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 7 |
Hb_000212_120 |
0.0876028653 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 8 |
Hb_005214_140 |
0.087686108 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 9 |
Hb_024184_010 |
0.0877831798 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 10 |
Hb_000300_270 |
0.0881118475 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_001876_040 |
0.0883297217 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 12 |
Hb_003417_040 |
0.0916092148 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 13 |
Hb_000230_130 |
0.0916585957 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006663_060 |
0.0917877664 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 15 |
Hb_022250_010 |
0.0921982772 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 16 |
Hb_004531_150 |
0.0924492904 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003040_050 |
0.095184786 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 18 |
Hb_002400_320 |
0.0954350835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_133702_020 |
0.0963152268 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_007597_010 |
0.096668864 |
- |
- |
hypothetical protein B456_010G072300 [Gossypium raimondii] |