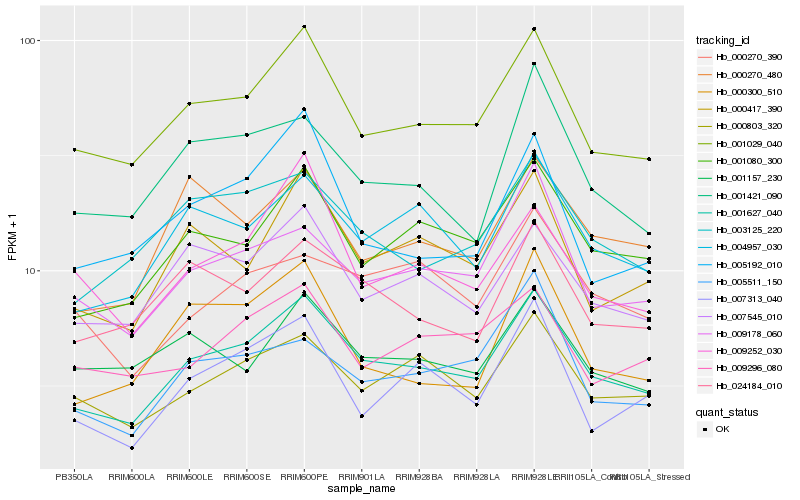
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001627_040 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001029_040 |
0.062554175 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 3 |
Hb_001080_300 |
0.0771777683 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 4 |
Hb_004957_030 |
0.0815775207 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 5 |
Hb_007545_010 |
0.0880379857 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 6 |
Hb_000803_320 |
0.0885601578 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 7 |
Hb_000417_390 |
0.0891935199 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 8 |
Hb_003125_220 |
0.0931982726 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 9 |
Hb_009178_060 |
0.0956384049 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At4g31390, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_024184_010 |
0.0974505705 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 11 |
Hb_001421_090 |
0.1029325417 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 12 |
Hb_000270_480 |
0.1036236506 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 13 |
Hb_007313_040 |
0.108650597 |
- |
- |
hypothetical protein JCGZ_23401 [Jatropha curcas] |
| 14 |
Hb_005192_010 |
0.1102860724 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_009252_030 |
0.1104739074 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 16 |
Hb_001157_230 |
0.1105477807 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 17 |
Hb_000270_390 |
0.1106815694 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 18 |
Hb_009296_080 |
0.1115704704 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000300_510 |
0.1118436632 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 20 |
Hb_005511_150 |
0.1118677 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |