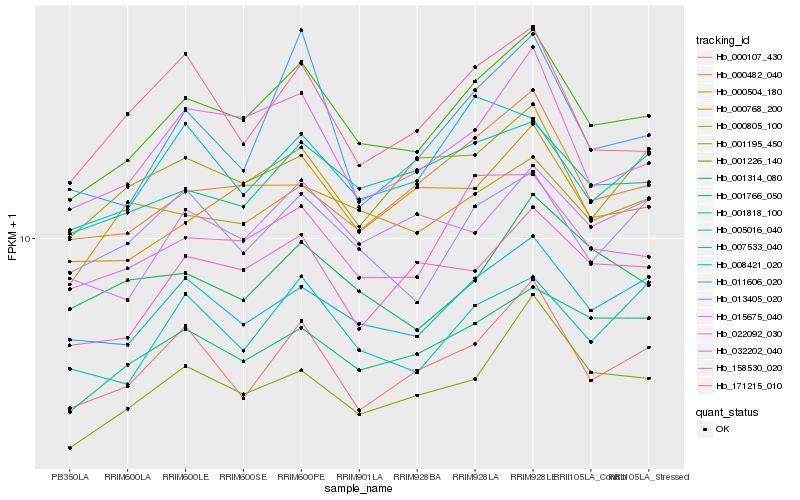
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001226_140 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 2 |
Hb_032202_040 |
0.0705528285 |
- |
- |
PREDICTED: phospholipase D beta 2 [Jatropha curcas] |
| 3 |
Hb_022092_030 |
0.0721525295 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_008421_020 |
0.0736066124 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000504_180 |
0.0824154315 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 6 |
Hb_001818_100 |
0.0826313208 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 7 |
Hb_158530_020 |
0.0836400187 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 8 |
Hb_000107_430 |
0.085336355 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 9 |
Hb_001766_050 |
0.0878458337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29290 [Jatropha curcas] |
| 10 |
Hb_001195_450 |
0.0881055818 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 11 |
Hb_000482_040 |
0.0898323574 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 12 |
Hb_000768_200 |
0.0924044301 |
- |
- |
hypothetical protein POPTR_0013s06910g [Populus trichocarpa] |
| 13 |
Hb_171215_010 |
0.0936971613 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 14 |
Hb_013405_020 |
0.0943216562 |
- |
- |
- |
| 15 |
Hb_015675_040 |
0.0946980338 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 16 |
Hb_000805_100 |
0.0955410898 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 10, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_005016_040 |
0.0960728969 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 18 |
Hb_011606_020 |
0.0975013437 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 19 |
Hb_001314_080 |
0.0978202427 |
- |
- |
hypothetical protein POPTR_0018s14330g [Populus trichocarpa] |
| 20 |
Hb_007533_040 |
0.0979643348 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |