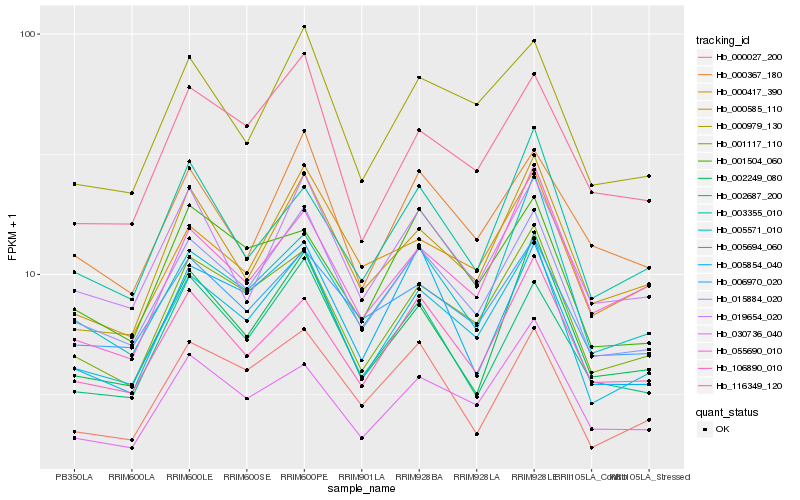
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_060 |
0.0 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 2 |
Hb_015884_020 |
0.041817512 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 3 |
Hb_001117_110 |
0.0470459853 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 4 |
Hb_000979_130 |
0.0659486831 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_000585_110 |
0.0778794886 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 6 |
Hb_030736_040 |
0.0789747306 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 7 |
Hb_116349_120 |
0.079725521 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_002249_080 |
0.080707961 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 9 |
Hb_106890_010 |
0.0809303733 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_005571_010 |
0.0852500336 |
- |
- |
PREDICTED: 15-cis-phytoene desaturase, chloroplastic/chromoplastic [Jatropha curcas] |
| 11 |
Hb_019654_020 |
0.0857310486 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 12 |
Hb_000027_200 |
0.087174948 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 13 |
Hb_006970_020 |
0.0874396815 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 14 |
Hb_000417_390 |
0.0896652301 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 15 |
Hb_055690_010 |
0.091177578 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001504_060 |
0.0914096545 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 17 |
Hb_003355_010 |
0.0914491042 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 18 |
Hb_002687_200 |
0.0930270352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000367_180 |
0.0946714059 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 20 |
Hb_005854_040 |
0.0951911844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |