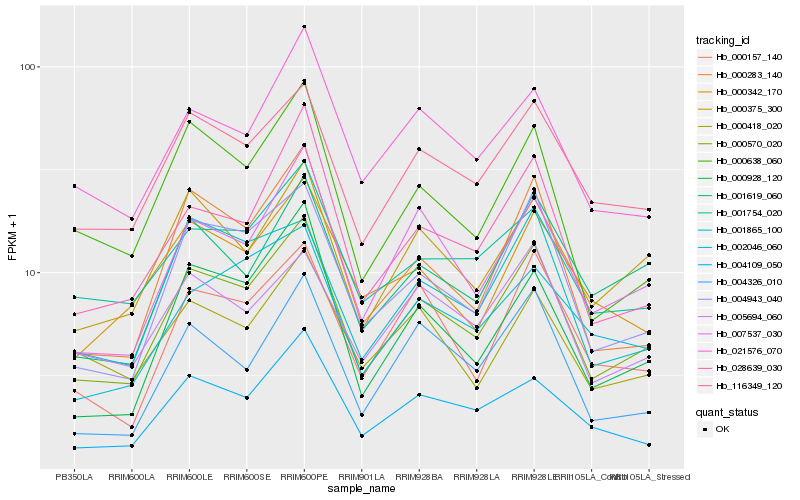
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000570_020 |
0.0 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 2 |
Hb_004943_040 |
0.07124931 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 3 |
Hb_116349_120 |
0.0802590244 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_007537_030 |
0.082154875 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000375_300 |
0.0894332372 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 6 |
Hb_000283_140 |
0.0895955439 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_028639_030 |
0.0923242431 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 8 |
Hb_004109_050 |
0.0928874702 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 9 |
Hb_000638_060 |
0.09537569 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 10 |
Hb_000157_140 |
0.0962185625 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 11 |
Hb_001754_020 |
0.0974593807 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000928_120 |
0.0977648816 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_021576_070 |
0.0979765519 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 14 |
Hb_004326_010 |
0.0999644 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_001619_060 |
0.1006588416 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005694_060 |
0.1028649174 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 17 |
Hb_001865_100 |
0.1031941198 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 18 |
Hb_000342_170 |
0.10785597 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 19 |
Hb_002046_060 |
0.1097118812 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 20 |
Hb_000418_020 |
0.1110306917 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |