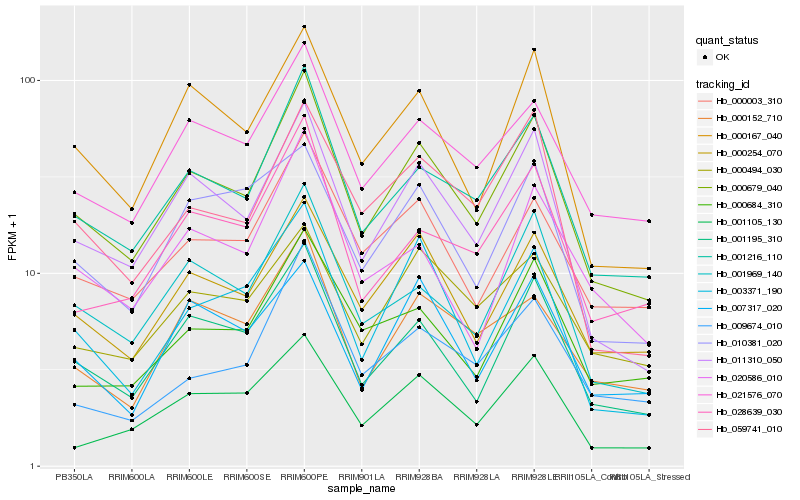
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_040 |
0.0 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 2 |
Hb_001216_110 |
0.058185863 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 3 |
Hb_011310_050 |
0.0688201416 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 4 |
Hb_001195_310 |
0.0730456457 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_001969_140 |
0.0855708728 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 6 |
Hb_000254_070 |
0.0861364477 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 7 |
Hb_021576_070 |
0.0885726911 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 8 |
Hb_000167_040 |
0.0887600274 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 9 |
Hb_000003_310 |
0.095774508 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 10 |
Hb_028639_030 |
0.104508146 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 11 |
Hb_000152_710 |
0.1065790365 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 12 |
Hb_001105_130 |
0.1072184301 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 13 |
Hb_059741_010 |
0.1078966524 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 14 |
Hb_000684_310 |
0.10839585 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 15 |
Hb_003371_190 |
0.1090074733 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 16 |
Hb_020586_010 |
0.1141172188 |
- |
- |
Alpha-N-arabinofuranosidase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000494_030 |
0.1171122025 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 18 |
Hb_009674_010 |
0.1186082182 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 19 |
Hb_010381_020 |
0.1204645918 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 20 |
Hb_007317_020 |
0.1206133537 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |